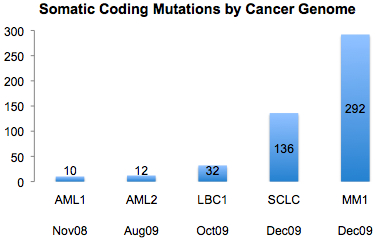
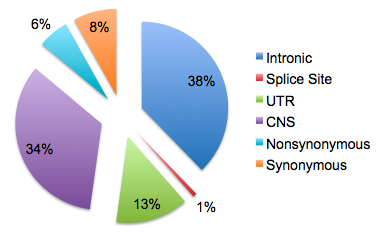
A recent article on GenomeWeb profiling the XGen Congress meeting in San Diego, where researchers debated the question of whether sequencing cancer genomes has clinical relevance. In a roundtable discussion, University of Washington’s Larry Loeb argued that cancer is too heterogeneous for sequencing to uncover the therapeutically-relevant mutations. As an example, he pointed to AML1 – the first cancer genome, which was sequenced here – whose 8 somatic mutations were not found in a screen of 187 other AMLs.
Recurrent Mutations in AML2
I wasn’t at the roundtable discussion, so I don’t know if anyone mentioned AML2 – the second cancer genome which was published in the New England Journal of Medicine – in which four of 64 somatic mutations were recurrent in other AMLs. Two of the recurrent mutations (NRAS and NPM1) were previously known. The other two were novel, and particularly interesting: one implicated IDH1 (a gene mutated in brain cancer) for the first time in AML, and the other was a noncoding (i.e. not-in-a-gene) mutation in an evolutionarily conserved region. The latter discovery reinforces the idea that “exome” sequencing – which targets the coding regions of known genes – can miss important variation, but that’s a debate for another day.
IDH1 and Clinical Relevance
I mentioned IDH1, which was first implicated in glioblastoma in a study led by Bert Vogelstein’s group at Johns Hopkins. In a Cancer Cell publication this January, members of the Cancer Genome Atlas (TCGA) research consortium found that abnormalities in IDH1 and three other genes (PDGFRA, EGFR, and NF1) helped distinguish clinically relevant subtypes in glioblastoma. Mutations in IDH1 have since been identified in other cancer types, including acute myelogenous leukemia (AML).
IDH1 encodes isocitrate dehydrogenase 1, a cytosolic enzyme that converts isocitrate to  -ketoglutarate. The discovery of somatic mutations in IDH1 in human cancers has put a spotlight on this gene and its mitochondrial homolog, IDH2. Recent studies have shown that mutations in IDH1 and IDH2 not only disrupt its normal activity, but create a new one: the reduction of
-ketoglutarate. The discovery of somatic mutations in IDH1 in human cancers has put a spotlight on this gene and its mitochondrial homolog, IDH2. Recent studies have shown that mutations in IDH1 and IDH2 not only disrupt its normal activity, but create a new one: the reduction of  -ketoglutarate to 2-hydroxyglutarate. Tumors with IDH1/IDH2 mutations show elevated levels of 2-hydroxyglutarate, suggesting that further studies of this “oncometabolite” may shed new light on cancer development and progression.
-ketoglutarate to 2-hydroxyglutarate. Tumors with IDH1/IDH2 mutations show elevated levels of 2-hydroxyglutarate, suggesting that further studies of this “oncometabolite” may shed new light on cancer development and progression.
If that’s not clinically relevant, I don’t know what is.
References
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, & Cancer Genome Atlas Research Network (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell, 17 (1), 98-110 PMID: 20129251