Myelodysplastic syndrome (MDS, also called preleukemia) is a blood disorder characterized by ineffective production of myeloid cells, or leukocytes. The disorderly and ineffective production of blood cells from stem cells in the bone marrow results in low blood counts, or cytopenias. As many of 30% of MDS cases progress to full-blown, chemotherapy-resistant secondary AML. This week in Nature Genetics, two studies report recurrent mutations in splicing-related genes in blood tumors.
First, Tim Graubert and colleagues describe the whole-genome sequencing of an MDS-derived secondary AML tumor and a matched normal (skin) sample. They detected and validated 507 somatic single-nucleotide variants in the tumor, nearly all of which (505) were detected in the MDS sample. Among these were 30 coding SNVs, of which one was a missense mutation in the U2AF1 gene. The same codon of U2AF1 was also mutated in two other MDS cases evaluated by WGS, highlighting it as a potential recurrently mutated gene. The authors undertook systematic sequencing of U2AF1 exons in 150 MDS cases, and found that 8.7% had mutations at Ser34.
Characterization of Recurrent U2AF1 Mutations
The authors undertook deep genomic resequencing, cDNA sequencing, and other experiments to characterize the nature of the U2AF1 mutations, finding that:
- Mutant allele frequencies were at 40-50%, suggesting that the mutation was present in most or all tumor cells.
- • SNP arrays and WGS indicated no large deletions or uniparental disomy spanning the U2AF1 locus.
- • Deep cDNA sequencing demonstrated that both wild-type and mutant alleles were expressed.
- • There were no apparent differences in the amount of U2AF1 mRNA between wild-type and mutated samples.
- • In the 150 cases examined, no other positions in the gene were mutated (other than residue 34).
Taken together, these observations suggest that U2AF1 alteration was an early, initiating event and likely represents a gain-of-function mutation.
U2AF1 and Splicing Factors
U2AF1 encodes a small regulatory subunit of the U2AF splicing factor. It binds the 3′ AG splice acceptor dinucleotide of the pre-mRNA target intron, and forms a heterodimer with U2AF2, which binds the adjacent polypyrimidine tract. U2AF1 is highly conserved, and loss of both copies is lethal in many species. Although it’s not known which domain of U2AF1 binds the mRNA, the Ser34 mutation occurs in a zinc-finger motif that may have RNA binding activity. Interestingly, in vitro reporter assays revealed that the Ser34 mutation causes an increase in splicing activity and more exon skipping relative to wild-type U2AF1. Further, an analysis of differentially expressed genes (by microarray) between samples with or without U2AF1 mutations revealed that three of the top functional categories for down-regulated genes were splicing- or RNA-recognition-motif-related genes. This observation may reflect one or more compensatory mutations for the increased splicing activity of U2AF1 mutants.
Recurrent Mutation of SF3B1 in Chronic Lymphocytic Leukemia
A second study in Nature Genetics, led by Victor Quesada and colleagues, employed exome sequencing to identify recurrent mutations in chronic lymphocytic leukemia (CLL), the most common form of adult leukemia in western nations. The authors sequenced the exomes of tumor samples and matched controls from 105 patients with CLL, 60 of which had mutated IGHV regions (a common alteration in CLL) and 45 of which did not. They reported ~45 somatic mutations per case, and observed more protein-altering mutations in IGHV-mutated samples (12.8 +/- 0.7) than non-IGHV-mutated (10.6 +/- 0.7). Comparing this study to their previous work (WGS of 4 CLL cases), the authors identified several new recurrently-mutated genes, including:
- • SF3B1, a subunit of the spliceosomal U2 snRNP11;
- • POT1, a nuclear protein involved in telomere maintenance12;
- • CHD2, which regulates gene expression by modification of chromatin structure13
- • LRP1B, which has recently been defined as a tumor suppressor in different malignancies
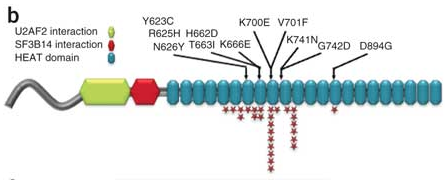
The authors focused on SF3B1 mutations, which was altered by somatic point mutations in ~10% of cases. Systematic screening of 279 cases by 3730 sequencing revealed that 9.7% of CLL tumors harbored SF3B1 mutations, making this the most frequently mutated gene in CLL identified to date. The protein encoded by SF3B1 is involved in the binding of the U2 snRNP to the branch point near 3′ splice sites. It interacts with RNA sequences and at least two proteins near the branch point: the early 3′-splice-site recognition factor U2AF65 and the branch point–binding protein SF3B14, as well as the RNA sequences near the branch point.
RNA-seq of SF3B1-mutated cases revealed some patterns of aberrant splicing, most of which paired a known 5′ donor site with a new, abnormal 3′ acceptor site. An analysis of splicing target genes revealed truncated versions of SLC23A2, a vitamin C transporter, and TCIRG1, one of whose gene products is a T-cell immune regulator. Another altered gene was FOXP1, known to be dysregulated in diffuse large B-cell lymphoma; the altered transcript lacked two PEST sequences normally required for protein degradation.
Role of Splicing in Tumor Development and Progression
Most adult tumors harbor hundreds or thousands of somatic mutations, only a fraction of which are likely to drive development and growth. Recurrence of mutations in the same gene or pathway remains the best way to isolate these “driver” mutations from background passenger events. These two studies, and a handful of others published late this year, suggest an important role for aberrant splicing in the early development of myeloproliferative disorders, such as MDS/sAML and CLL. What’s particularly important is that these appear to be gain-of-function mutations, which opens the door to new potential targeted therapies. It’s one step closer to personalized medicine for cancer patients, brought to you by next-generation sequencing.
References
Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, McLellan MD, Dooling DJ, Abbott RM, Fulton RS, Schmidt H, Kalicki-Veizer J, O’Laughlin M, Grillot M, Baty J, Heath S, Frater JL, Nasim T, Link DC, Tomasson MH, Westervelt P, Dipersio JF, Mardis ER, Ley TJ, Wilson RK, & Walter MJ (2011). Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nature genetics PMID: 22158538
Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, López-Guerra M, Colomer D, Navarro A, Baumann T, Aymerich M, Rozman M, Delgado J, Giné E, Hernández JM, González-Díaz M, Puente DA, Velasco G, Freije JM, Tubío JM, Royo R, Gelpí JL, Orozco M, Pisano DG, Zamora J, Vázquez M, Valencia A, Himmelbauer H, Bayés M, Heath S, Gut M, Gut I, Estivill X, López-Guillermo A, Puente XS, Campo E, & López-Otín C (2011). Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature genetics PMID: 22158541