Two studies published in JAMA reveal the power of whole-genome sequencing improve the diagnosis and treatment of cancer. Both are from collaborations of the Genome Institute and the School of Medicine at Washington University in St. Louis, Missouri. First, Link and colleagues report the identification of a novel cancer susceptibility mutation (a 3-kbp deletion in TP53) by whole-genome sequencing of a patient with therapy-related AML (tAML). In the same issue, Welch et al use whole-genome sequencing to diagnose a cryptic fusion oncogene (bcr3 PML-RARA) in a patient with acute promyelocytic leukemia (APL).
Early-onset Cancer and Therapy-related Leukemia
The first story, unfortunately, is a sad one: the patient, a 37-year-old woman, first presented with stage II ER+/Her2+ breast cancer. She underwent surgery, radiation, and chemotherapy. Two years later, she was diagnosed with stage IIIC ovarian carcinoma, and again had surgery and chemotherapy. At age 42, the ovarian cancer was back, and six months later, the patient presented with t-AML. She developed respiratory failure and died within days.
Therapy-related leukemia is a rare but well-documented complication among cancer patients, and thought to be a direct consequence of the cytotoxic effects induced by chemo and/or radiation. Classic tAML usually presents with “unfavorable cytogenetics” – in other words, large-scale structural changes to chromosomes that are visible under a microscope. The t-AML studied here was thus very different from the first two published cancer genomes (AML1 and AML2), which were cytogenetically normal. Indeed, cytogenetic analysis of t-AML1 revealed a complex karyotype:
- Monosomy-7, meaning that only one copy of chromosome 7 was present, when there should normally be two
- 5q(del), in which the long arm of chromosome 5 has been lost. This is common in tAML
- 2 marker chromosomes that could not be identified
The t-AML might be expected, given the patient’s multiple rounds of chemotherapy and radiation. Yet her early-onset diagnoses of breast and ovarian cancer are a bit puzzling. None of her first-degree relatives had had cancer, and commercial tests for BRCA1/BRCA2 mutations came back negative.
Revelations of Whole-Genome Sequencing
To identify susceptibility variants and somatic mutations in the patient’s leukemia, Link and colleagues performed whole-genome sequencing on DNA from the tumor (blood) and matched normal tissue (skin). High-density SNP array and spectral karyotyping data were also generated, to help resolved the complex cytogenetic alterations. SV and copy number analysis of the WGS data confirmed the monosomy-7 and 5q loss, and also identified several smaller alterations not visible to karyotyping. Altogether, the authors identified and validated 28 somatic coding mutations (26 point mutations, 2 small indels), 8 somatic structural variants, and 12 acquired copy number alterations (CNAs). None of the genes harboring SNVs or small indels were mutated in a panel of 93 other t-AML samples, so it’s difficult to determine which of them contributed to tumorigenesis.
Identification of a Deleterious TP53 Deletion
There was one particularly interesting finding: a homozygous 3-kb deletion of TP53. Sequence data enabled precise localization of the breakpoints of the deletion, revealing that it removed exons 7 to 9 of the gene, effectively ablating the DNA-binding domain of the p53 protein. Clearly, a deleterious mutatation, and almost certainly the key susceptibility mutation that explains the early-onset breast and ovarian cancer. Strikingly, the deletion was present in heterozygous form in the normal skin DNA. The patient’s mother did not carry it, and while her father was no longer alive, his extended family had no history of cancer, making it unlikely that he carried it.
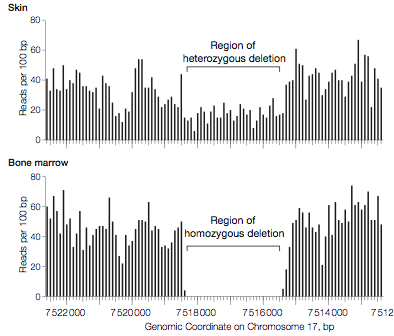
Credit: Link et al, JAMA 2011
The authors conclude that the 3-kbp deletion was a de novo germline mutation. That is, the mutation occurred spontaneously; the patient was born with it, but her parents didn’t have it. Recent whole-genome sequencing studies of mother-father-child trios have shown that de novo mutations do occur in human generations, albeit at a very low rate. It’s a matter of unlucky chance to acquire a mutation in a key tumor suppressor gene. All it takes is a single somatic mutation in the functional copy, or loss of the chromosome, to get a cell with no functional p53. In the leukemic cells, for example, there was a uniparental disomy event (two copies of a chromosome from one parent, instead of one from each parent) of chromosome 17, rendering the deletion homozygous. A double-knockout of TP53.
Implications and Genetic Counseling
The presence of the deletion in skin DNA was worrisome, because it suggests that this was part of the patient’s overall genetic makeup. She had children. Would they have inherited the mutation as well? Fortunately, anticipating that they might find clinically-relevant information, Link and colleagues wrote into their consent protocols a mechanism to communicate such information back to the family. They contacted the primary care physician, who discussed the issue with the patient’s family members, and encouraged them to seek genetic counseling. So, if I can offer a silver lining: even though the patient died, her contribution to this study yielded new knowledge that might one day save the lives of her children.
Diagnosis of a Cryptic Fusion Oncogene
The second study, by Welch and colleagues, is hands-down one of the most compelling success stories of whole-genome sequencing to date. The patient was a 39-year-old female with acute leukemia. She’d begun standard “induction” chemotherapy (as in “induce remission”) with cytarabine, idarubicin, and all-trans retoinoic acid (ATRA). However, her metaphase cytogenetics revealed a complex pattern of deletions and rearrangements, which is associated with poor (15%) long-term survival and thus, treated by allogenic transplantation during first remission whenever possible. At this point, she was referred to the authors’ institution. Fluorescent in-situ hybridization (FISH), a more sensitive test, suggested a RARA-PML fusion (not PML-RARA).
At this point, there was a “diagnostic conundrum” and ATRA was discontinued. At day 14, she had persistent AML – the standard chemotherapy sans ATRA wasn’t working. The question was this: did she have unfavorable-prognosis, cytogenetically-complex AML, or did she have AML with promyelocytic features (APL)? If it was the former, she needed a transplant – an expensive, painful operation with various other risks.
In contrast, PML tends to have a favorable prognosis. It’s commonly (90%) associated with PML-RARA fusion transcripts that arise from a t(15;17) translocation. The good news is that such patients usually respond to all-trans retinoic acid (ATRA), because the RARA gene encodes a retinoic acid receptor. Indeed, treatment with ATRA leads to substantially improved outcomes (69% 5-year disease-free survival, compared to 29% for standard chemotherapy alone). But for the patient in question, ATRA had been discontinued because the classic fusion (PML-RARA) wasn’t there.
She was consented for whole-genome sequencing, which was performed using Illumina paired-end reads on tumor (blood) and matched normal (skin) DNA. This time, however, the researchers didn’t have the luxury of months or years to complete the sequencing and analysis – her doctors were waiting on the results for a treatment decision. How quickly could it be done, and a decision reached? Here was the actual timeline:
| Day | Status |
| 1 | Sample collected |
| 5 | Libraries made, sequencing begins |
| 18 | Sequencing complete |
| 22 | Reference alignment complete |
| 24 | SNVs called by SomaticSniper |
| 25 | Structural variants called by BreakDancer |
| 52 | Fusion events validated, physician informed |
Whole-genome sequencing identified 12 valid somatic SNVs (point mutations) of unknown significance, whose allele frequencies suggested two distinct leukemia clones in the bone marrow. WGS identified all of the cytogenetically-visible rearrangements as well as two other large deletions (on chromosomes 14 and 19) that were missed by cytogenetics. It also revealed that apparent losses of chromosomes 6 and 16 were incomplete; instead, these chromosomes were partially represented on two unidentifiable “marker” chromosomes.
Resolution of a Cryptic Rearrangement of PML and RARA
Finally, and most importantly, the sequencing enabled breakpoint detection and assembly of a complex rearrangement of three genes: LOXL1 and PML on chromosome 15, and RARA on chromosome 17. In addition to the translocation, a 77-kb insertion and a 77-kb deletion had created two “derivative” chromosomes. That’s what had confounded the FISH analysis.
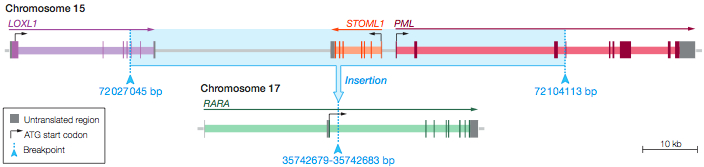
Credit: Welch et al., JAMA 2011
PCR confirmed that three fusion transcripts were present: bcr3 PML-RARA, LOXL1-PML, and RARA-LOXL1. Expression of PML-RARA was confirmed by RT-PCR. The detection of a PML-RARA fusion supersedes cytogenetic findings, and gave the patient a favorable prognosis. She was given ATRA, and at the time of writing, had been in complete remission (no detectable leukemia) for 15 months and counting.
Whole-genome sequencing is not ready for widespread clinical application just yet. The cost at the time was roughly $80,000 for the two samples (tumor and normal), and it’s not covered by insurance. Validation, a critical step, remains a bottleneck. In spite of these considerations, the authors have a major accomplishment on their hands. Whole-genome sequencing, analysis, and validation were finished within 2 months. It resolved the complex PML-RARA event, and did so in time to make a life-saving treatment decision.
References
Link DC, Schuettpelz LG, Shen D, Wang J, Walter MJ, Kulkarni S, Payton JE, Ivanovich J, Goodfellow PJ, Le Beau M, Koboldt DC, Dooling DJ, Fulton RS, Bender RH, Fulton LL, Delehaunty KD, Fronick CC, Appelbaum EL, Schmidt H, Abbott R, O’Laughlin M, Chen K, McLellan MD, Varghese N, Nagarajan R, Heath S, Graubert TA, Ding L, Ley TJ, Zambetti GP, Wilson RK, & Mardis ER (2011). Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA : the journal of the American Medical Association, 305 (15), 1568-76 PMID: 21505135
Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, Wallis J, Chen K, Payton JE, Fulton RS, Veizer J, Schmidt H, Vickery TL, Heath S, Watson MA, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Ley TJ, & Wilson RK (2011). Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA : the journal of the American Medical Association, 305 (15), 1577-84 PMID: 21505136
[…] science blogs, etc. are strange places to find interesting literature, but they work for me. This April 22 post on the wonderful Massgenomics blog alerted me to two interesting papers published in JAMA on April […]