This week in Nature, investigators from Wellcome Trust Sanger Institute published the fourth and fifth complete cancer genomes. Interestingly, both are cancers in which the primary mutagen is known: malignant melanoma (UV light) and small-cell lung cancer (tobacco smoke). This seems to be important, because when I looked at the number of validated somatic coding mutations in each of the first five genomes, the latest two stood out.
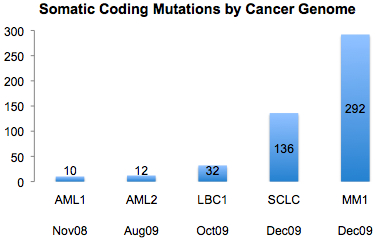
Granted, small-cell lung cancer and malignant melanoma differ in many ways from leukemia and breast cancer. Yet the increase of confirmed somatic coding variants in these two recent studies is striking.
Corregendum: Not the First Catalogue of Somatic Mutations
Even so, the authors’ claim that they are “providing the first comprehensive catalogue of somatic mutations from an individual cancer” seems unjustified. Perhaps this is based on the idea that AML1 and LBC1 focused on coding variants only. Yet I point out that in AML2 we evaluated 282 noncoding somatic mutations and confirmed over 50. In the melanoma study, only 470 of the 33,345 newly found somatic mutations were sent through validation, and the method for selecting these was not made clear. At best, the “first comprehensive” claim is a semantic one; at worst, it’s just wrong.
That said, these are still landmark studies. Even with the emergence of next-generation sequencing, completing a cancer genome is a marvelous achievement. It requires substantial financial resources and technical expertise; we certainly knew that WTSI had these. But guiding the data through analysis and forming a cohesive story out of it is the real challenge. It requires persistence, intellect, and scientific rigor, but most of all it requires strong leadership. I congratulate our friends across the pond for showing that they have what it takes.
Illumina’s Fourth, ABI SOLiD’s First Cancer Genome
The melanoma study, like the first three cancer genomes, applied high-throughput sequencing on the Illumina platform (2 x 75bp, in this case). In contrast, the SCLC study is the first cancer genome to be sequenced on a different platform – ABI SOLiD. While the read lengths for SOLiD were not impressive (2×25 bp), the specificity was – 77 of 79 (97%) of somatic coding SNVs and 333 of 354 (94%) randomly chosen genome-wide variants tested confirmed by PCR and traditional sequencing.
Unfortunately, SOLiD also comes with a sensitivity cost. Only 22 of 29 previously identified SNVs (77%) were called. Indels were a real problem – neither of the two previously known coding indels were detected, and the validation rate for predicted somatic indels was 25%.
Mutational Signatures Implicate UV Light and Tobacco
Intriguingly, in both studies the authors identified distinct mutational signatures of exposure to the long-suspected environmental risk factor – ultraviolet radiation (in malignant skin cancer) and tobacco smoke’s “cocktail of carcinogens” (in lung cancer). The substantial number of mutations in each genome made it possible to characterize these signatures with unprecedented statistical power.
The strength of both studies is the insight into the molecular mechanisms of DNA damage, repair, and mutation that could be inferred from such a powerful dataset. In melanoma, the most prevalent mutations were C->T and G->A transitions; the mutational spectrum and sequence context indicate that most of these are attributable to ultraviolet-light-induced DNA damage. In lung cancer, G->T transversions were the commonest substitution; these mutations have previously been linked to polycyclic aromatic hydrocarbons and acrolein in tobacco smoke.
Don’t Smoke. Wear Sunscreen.
Cancer, like many common, complex diseases, has many risk factors that come from the environment as well as from one’s DNA. Here, for perhaps the first time, we get a picture of the significant mutational burden associated with two lifestyle choices. Smoking is an obvious one – avoiding it dramatically decreases one’s risk of cancer and a host of other diseases. Now science can offer another definitive piece of advice. Everybody’s free, as Baz Luhrmann puts it, to wear sunscreen.
References
Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, & Stratton MR (2009). A comprehensive catalogue of somatic mutations from a human cancer genome. Nature PMID: 20016485
Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, Varela I, Nik-Zainal S, Davies HR, Ordoñez GR, Mudie LJ, Latimer C, Edkins S, Stebbings L, Chen L, Jia M, Leroy C, Marshall J, Menzies A, Butler A, Teague JW, Mangion J, Sun YA, McLaughlin SF, Peckham HE, Tsung EF, Costa GL, Lee CC, Minna JD, Gazdar A, Birney E, Rhodes MD, McKernan KJ, Stratton MR, Futreal PA, & Campbell PJ (2009). A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature PMID: 20016488

