Micro-RNAs (miRNAs) are short (18-26 nt) sequences that act as post-transcriptional repressors of gene expression. Over 700 miRNAs have been reported in the human genome; each is believed to bind directly to many mRNAs to regulate their translation or stability. Thus, miRNAs represent a key regulatory mechanism affecting numerous cellular activities, and are of particular interest in cancer research. Understanding the complex relationships between miRNAs and mRNAs remains challenging, however, and computational approaches alone have been largely unsuccessful.
HITS-CLIP: Isolation and Sequencing of Argonaute-miRNA-mRNA Complexes
Enter HITS-CLIP, a new approach that applies high throughput sequencing of RNAs isolated by crosslinking immunoprecipitation. Essentially, it’s a method by which radition is used to cross-link protein-RNA complexes and stringently purify them. Then, massively parallel sequencing yields all of the RNA “tags” bound by the protein of interest.
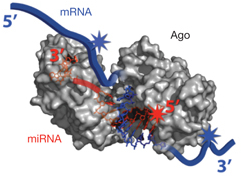
Ago-miRNA-mRNA Complex (Image Credit: Nature 460: 479-486, 2009)
In a recent Nature paper, Chi et al used HITS-CLIP to isolate RNA bound by the Argonaute protein (Ago), which mediates miRNA-mRNA interaction (see figure). The purified complexes showed two different modal sizes (110 kDa and 130kDa), suggesting that Ago (97 kDa) was crosslinked to two different RNA species – hopefully, miRNAs (small) and the mRNAs that they were targeting (large).
The authors applied Illumina high-throughput sequencing to characterize Ago-bound miRNAs and the mRNA “tags” to which they were linked. With relatively straightforward bio-informatics approaches, it was possible to cross-reference expressed miRNAs with complementary sequences of mRNA tags. The resulting “ternary map” of miRNA-mRNA interaction sites yields a wealth of information about this post-transcriptional regulatory mechanism.
Decoding miRNA-mRNA Interaction
The authors identified 454 unique miRNAs crosslinked to Ago in the mouse brain; mir-30e was the most abundant species, representing 14% of all miRNA tags. In silico clustering and normalization of the messenger RNA tags yielded 1,463 robust clusters from 829 different brain transcripts.
Locations of Ago-bound mRNA tags (Image Credit: Nature 460: 479-486, 2009)
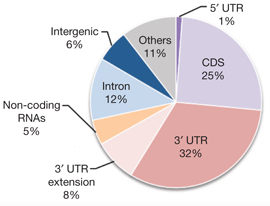
When these tags were overlaid with gene annotations, several patterns emerged. As expected, a substantial portion (40%) of Ago-bound tags were in 3′ UTRs where miRNA activity is known to have high efficacy. Some 8% (one-fifth of the 40%) were actually outside of the UTR but <10kb downstream, regions likely to harbor unannotated 3′ UTRs.
Unsurprisingly, very few Ago-bound tags were in 5′ UTRs. However, a substantial fraction of tags fell in coding sequences (25%), introns (12%), and non-coding RNAs (4%), suggesting that miRNA activity occurs in these regions as well. Another 6% of tags were in intergenic regions, possibly in as-yet-unannotated transcripts. These unexpected locations of miRNA binding may offer additional insights into the mechanisms of miRNA regulation.
Next, the authors sought to define the Ago-mRNA “footprint” in which the majority of tags were contained. The distribution of tags in a defined cluster, at least in their figure, looks like a bell curve, with a sharp peak in the middle. About 95% of the time, Ago bound within 45-62 nucleotides of this peak, so the authors defined this region as the average Ago-miRNA footprint. Linear regression analysis of all 6-8 base motifs in clusters yielded numerous “enriched” seed sequences; the most prevalent corresponded to the binding site of miR-124, a well known brain-specific miRNA. Indeed, Ago-mRNA footprints were rich in miRNA binding sites, suggesting that this approach may predict active sites with far better specificity than other methods.
HITS-CLIP Implications
By reducing the search space for miRNA binding sites to a 45-60-nucleotide Ago footprint, HITS-CLIP offers a powerful complementary approach to bioinformatic methods for miRNA binding site prediction. Computational approaches alone are known to have high false positive rates, whereas the authors estimate FP rates of just 15% for HITS-CLIP. The new method offers dramatic improvement for transcripts with highly conserved 3′ UTRs, which often have many “predicted” miRNA binding sites because so many computational methods rely on conservation. Analysis of the HITS-CLIP ternary map revealed that real miRNA-mRNA binding events are very specific, with an average of just 2.6 Ago-mRNA clusters per regulated transcript. Despite the thousands of predicted binding sites, each miRNA bound an average of 655 targets. These results suggest that miRNA selectivity is much higher than previously believed. Yet Ago-mRNA clusters seemed to show no apparent sequence preference (data not shown), so it’s likely that other RNA-binding proteins are involved.
Thus, this study sets the stage for large scale genome-wide RNA-protein maps that include other proteins, tissues, and species, which should yield an unprecedented new level of understanding of this complex regulatory process.
References
Chi SW, Zang JB, Mele A, & Darnell RB (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature, 460 (7254), 479-86 PMID: 19536157