This month in Nature, a group from Jay Shendure’s lab reported perhaps the most ambitious targeted resequencing study to date – the whole exome sequences of 12 individuals.

Using an array-based hybridization capture method (2 microarrays, 10  g of input DNA), Ng et al selectively targeted CCDS regions totaling 26.6 Mb of sequence (~0.83% of the human genome). Capture specificity was similar to that of other published methods (35-55% of reads mapping to targets), but the completeness was astonishing – on average, 99.7% of target bases covered at least once and 96.3% covered at 8x with q>=30.
g of input DNA), Ng et al selectively targeted CCDS regions totaling 26.6 Mb of sequence (~0.83% of the human genome). Capture specificity was similar to that of other published methods (35-55% of reads mapping to targets), but the completeness was astonishing – on average, 99.7% of target bases covered at least once and 96.3% covered at 8x with q>=30.
By focusing on coding exons, the authors achieved 51x coverage (on average) with just 6.4 Gb of mappable sequence per individual. Illumina 76-bp single-end sequencing was the platform of choice. If I make some rough empirical estimates of mapping rate and reads per lane, they generated a single Illumina run of data (7-8 lanes) per individual. Compared to whole-genome sequencing, the authors claim a 20-fold reduction in the amount of sequence required. I’d say this estimate is pretty close. Our second leukemia genome, which had 23x haploid coverage, took 16.5 Illumina runs to complete.
Strong Illumina Pipeline
It’s not simply the technological feat that impressed me about this study. The presentation of the work and underlying analytical approaches are just outstanding. While reading through the methods, I couldn’t help but think that nearly every step the authors took in processing their data was something that we’ve implemented here – Maq alignment, start site de-duplication, mining Maq-unplaced reads for indels, etc. We have a bit of a friendly rivalry with University of Washington (since we are, after all, Washington University), so I looked for weak points. Try as I might, I couldn’t find much to criticize about the analysis. When it comes to Illumina sequencing, UW seems to know what they’re doing.
How to Write A Nature Paper
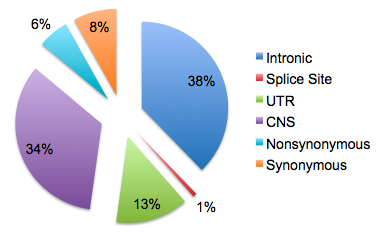
And paper itself is just clear, concise, well-written – everything I’d expect from a Nature publication. Take Figure 1, for example. Figure 1, in general, is the focal point of most research papers, and for that reason I think many authors try to cram way too much into it. Not this time. Four histograms that all have “Number of observations of minor allele” as their X-axis. Yet each one tells a different story: (a), how novel-to-dbSNP variants were rare; (b), how nonsynonymous variant frequencies are shifted to lower values relative to those of synonymous variants, (c), how this shift in allele frequencies is more pronounced for damaging nsSNPs, consistent with natural selection, and (d), how the sizes of observed indels are enriched for non-frameshift events divisible by 3.
Illumina Sequencing and Deduplication
Early into our days of Illumina/Solexa sequencing, we observed a strange phenomenon in the data: lots of reads with identical start sites and orientations. The theory was that these occasional pileups were PCR-related, and each one arose from a single molecule that somehow was sequenced over and over again. Since just about every downstream analysis (coverage, mutation detection, etc.) relies on unbiased read counts, it’s important to normalize for such events. This requires a “de-duplication” step in which multiple reads with the same start site and orientation (presumably the same molecule) are discarded and only one is kept.
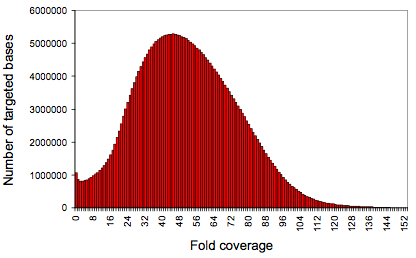
Credit: Nature 461:272-276 (2009)
The implications of this deduplication requirement, as pointed out by Ng et al, are that the maximum read depth for any given position in the genome is twice the read length for single-end libraries. In their case, 152x. One might be concerned that even with de-duplication there would be substantial bias in targeted capture. But look at the bell curve of the coverage distribution from supplemental figure 1 (left).
Someone had better call O’Reilly, because that’s just beautiful data. Importantly, the deduplication paradigm changes somewhat for paired-end sequencing, which is largely what we do here. With paired ends, you have two reads from each molecule, each with a start site and orientation. So the maximum coverage immediately jumps to 4 times the read length. Furthermore, due to the variation in fragment sizes of sheared DNA, insert sizes add further distinction for different molecules, allowing for read depths of 1000x or more after de-duplication for paired-end reads.
Identifying Disease-Causing Mutations
What pleased me most about this study is that the authors didn’t just present exome capture and sequencing of “undiseased” individuals. In addition to 8 HapMap samples, they included four samples from unrelated individuals with Freeman–Sheldon syndrome (FSS), an autosomal-dominant disorder caused by mutations in MYH3. After collecting the set of coding variants in each individual, the authors asked a simple question: could we have pinpointed the disease gene from mutation data? With the knowledge in hand that this was a monogenic, autosomal-dominant disorder, the authors assumed that the same gene might be mutated in most (or all) samples. And since the disease itself is uncommon, the authors inferred that common variants could be excluded. So, with the full set of mutations for each affected individual in hand, the authors looked for genes where:
- There was at least one (but not necessarily the same) nonsynonymous SNP, splice-site SNP, or coding indel in all four samples.
- The mutations were novel; that is, they weren’t found in dbSNP or the other 8 HapMap samples.
- The mutations were predicted to damage the encoded protein
When these criteria were applied, the authors whittled down a list of 4,510 genes with mutations in at least one sample to just 1, and that gene was MYH3. Thus, whole-exome sequencing allowed for direct identification of a disease-causing gene with just a few samples from affected individuals. Granted, the authors got lucky. The causal mutations might have been SVs, or missed by variant callers, or not covered sufficiently by sequence data. Or, the disorder might be caused by a single mutation in one of several genes, as is the case of autosomal dominant RP, a monogenic disorder for which at least 16 genes have been implicated.
Even so, the authors applied a relatively straightforward approach and got the right answer. With whole-exome sequencing capability within reach, finding the genes behind autosomal disorders is only a matter of time.
References
Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, & Shendure J (2009). Targeted capture and massively parallel sequencing of 12 human exomes. Nature, 461 (7261), 272-6 PMID: 19684571