As cancer genome sequencing ramps up here and pretty much everywhere around the world, I got to thinking about strategies for identifying somatic changes, with confidence, from massively parallel sequencing data. As part of the Cancer Genome Atlas Project (TCGA), we’ve been applying both targeted (capture-based) and whole-genome sequencing approaches to tumor samples and matched normal controls. Ideally, the resulting data will yield high (>20x) coverage in both tumor and normal across our positions of interest. What happens next, at least at WashU, is the culmination of a multiple-year effort to develop a comprehensive pipeline for detecting somatic variants.
First up: Single Nucleotide Variants (SNVs)
With more than 15 million entries in dbSNP, single nucleotide polymorphisms (SNPs) remain the most common form of DNA sequence variation in humans. In cancer, most of the well-characterized somatic mutations are single nucleotide changes as well. Conceptually, SNVs should be the easiest things to find in next-gen sequencing data. They occur at a single position that can be directly compared between tumor and normal. They should have minimal effects on sequence alignments to the reference genome. For example, here’s a putative somatic variant in TP53:
![]()
What you see above is SAMtools “pileup” output at a single position (7518990 on chr17), for Normal and Tumor. The Normal shows 4 reads that all support the reference on the – strand (,,,,). The Tumor, however, shows 6 reads that all support a G variant, 2 on the + strand (GG) and 4 on the – strand (gggg). It seems reasonable that, given this output across the entire genome for Normal and Tumor, one can compare them at every position and look for differences such as these.
Yet we struggle to validate even high-confidence SNVs that look to be somatic. Some are real, but Germline (probably under-sampled or missed in the Normal); most are simply false positives in the tumor. These might arise from a number of causes – homopolymers, paralogs, repeats, sequencing error, alignment error, etc. Only a small fraction of variants that appear somatic in NGS data will validate as such.
Why is that? In general, it’s because by screening for somatic variants, we remove all of the variants that are most likely to be real. First, we exclude any variants that are present in the normal (germline) – which account for the majority of true sequence variations. We also exclude known variants from dbSNP and 1,000 Genomes databases, which are also likely to be real but almost certainly germline events. Then, we prioritize variants that are predicted to have functional effects – on protein coding, on splicing, in conserved regions, etc. Such regions are often under negative selection for damaging mutations, meaning that variants should be exceedingly rare. Every one of these filters selects for variants that are less likely to be valid.
Small Indels
With longer (>50 bp) fragment-end reads and/or paired-end libraries, it’s possible to detect small insertion/deletion variants (indels) in next-gen sequencing data. Here, detection and specificity are the challenges. In 454 data, the reads are [hopefully] of sufficient length (250 bp) for accurate gapped alignment to a reference sequence, and indeed, aligners commonly used with 454 data (Newbler, BLAT, cross_match, SSAHA2) do so. Unfortunately, indels are both the strength and the weakness of 454 data – due to the underlying pyrosequencing, homopolymeric regions are often under- or over-called, resulting in numerous false positives. Many can be filtered, but often homopolymer-associated errors cause mis-alignment of reads, yielding indels that might not look like homopolymer artifacts.
Indel detection is also possible with Illumina data, though the shorter read lengths make this challenging. Few short read aligners can handle the throughput of Illumina data and allow for gaps in read alignments, because speed and gapped alignment are at odds with one another. Fortunately, paired-end sequencing on Illumina offers a solution implemented by Maq some time ago – first, map all reads that you can without gaps, and then, look for gapped alignments in unplaced reads whose mate is mapped nearby. This reduces the search space considerably for gapped alignment, and also limits the query space to reads that likely contain indels (gaps).
In cancer sequencing, small indels present one additional problem – determining whether they are present in the normal. Even the best aligners can’t always precisely define where an indel starts or stops. Thus, a germline indel might have different coordinates in the tumor than in its matched control; when comparing the samples, it might appear to be somatic.
Loss of Heterozygosity (LOH)
It is well known that the genomes of tumor show extensive loss of heterozygosity (LOH). Generally, this occurs because a position that is heterozygous in the germline is affected by some kind of structural event – deletion, gene conversion, chromosome loss, etc. – that results in the loss of one allele. Of course, to detect LOH, one needs a variant that’s heterozygous in the Normal, and to precisely define the region of LOH, one needs a dense set of heterozygotes. Even so, the maximum precision for the start and stop of an LOH region is the interSNP distance, since only SNPs can inform on LOH, and that can be hundreds or thousands of bases. But LOH calls do tend to cluster, and detection of LOH regions is not really the problem. Even lower-resolution array technologies identify recurrent LOH regions in tumor samples.
But what exactly does LOH mean in terms of cancer development and growth? It’s hard to say. Quite possibly, a tumor suppressor gene was deleted, or an oncogenic allele was duplicated. Unfortunately, LOH regions tend to be kilobases or megabases in size, containing dozens or hundreds of genes, and identifying which ones are truly affected in terms of cancer remains challenging. We see a lot of LOH in cancer, but sadly, it never seems to get anyone excited.
Structural and Copy Number Variation
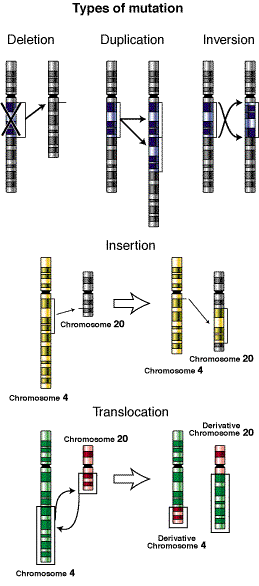
Image Credit: Wikipedia
Last and most difficult to characterize are the sub-microscopic structural changes – insertions, deletions, inversions, translocations, duplications, etc. – that often occur in tumor genomes. These tend to be large, complex events that are tough to infer from NGS data. We run Ken Chen’s breakDancer, of course, and it predicts numerous SVs. But how do you validate a massive, complex variant spanning thousands of bases? We do our best with PCR and 3730/454 sequencing, but until read lengths get really really long (perhaps on single-molecule sequencing), validating such events and determining their breakpoints is tough.
There are well-characterized, recurrent copy number alterations in cancer, like EGFR amplification on chromosome 7. Here’s my question: where are all of those extra copies? Are they just tandem duplications of part of a chromosome, or are they duplications that get inserted elsewhere in the genome? In the absence of a complete, linear, high-confidence genome, I’m not sure we can tell.
Fruits of Our Labors
It occurs to me that this is a bit of a negative article – focusing entirely on the challenges and failures, without highlighting the successes. And there are many successes. Every cancer genome tells us something, and every new piece of knowledge goes into our arsenal in the war against cancer. As sequencing ramps up, we’ll see exponential growth in the number of known somatic mutations across a wide array of cancers. With the help of cancer biologists, these data will be leveraged to better understand the genes, proteins, and pathways underlying tumorigenesis. Greater understanding will undoubtedly improve the detection, diagnosis, prognosis, and treatment of cancer patients.
Just warn you that maq’s way to detect short indels probably does not work well when reads become longer. Some preliminary analysis implies that using an aligner capable of gapped alignment for SE reads leads to significantly more true indel calls than using maq. In addition, when reads go longer, the chance of both ends having indels is also higher. Nonetheless, maq’s strategy is still useful in identifying a bit longer indels (>6bp) as few aligners allow long indels for SE reads for the sake of efficiency and alignment accuracy as well.