The cost of DNA sequencing continues to plummet, and while insurance companies might not be ready to get on board, another study has demonstrated how individual genome data can be clinically informative. Jonathan Rios and colleagues took on the case of an 11-month-old girl with severe hypercholesterolemia (1023 mg/dl) whose parents were unaffected, which suggested either an autosomal recessive disorder or a de novo mutation. A normal plasma sitosterol:cholesterol ratio ruled out sitosterolemia, and a screen of commonly-mutated hypercholesterolemia genes (LDLRAP, LDLR, PCSK9, APOE and APOB) came up empty.
Enter Whole-Genome Sequencing
Whole-genome resequencing for the patient was performed by Complete Genomics, who generated 138 Gbp of mappable sequence yielding 49x average coverage. Of the ~3.8 million sequence variants identified, 502,000 were indels or complex rearrangements that were not further evaluated (see Figure 2A). That left ~3.3 million SNPs, of which 9,726 were nonsynonymous or splice site variants. Most of these were present either in dbSNP or in the genomes of 21 unaffected individuals (16 exomes, 5 whole-genomes). About 700 variants remained.
Now the authors considered the disease pedigree:
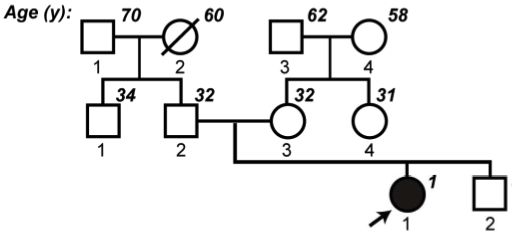
- Rios et al., Hum. Molec. Genetics 2010
The parents were unrelated (non-consanguinous) and both had normal cholesterol. No other relatives (including the patient’s 4-year-old brother) were affected. It fit with a monogenic autosomal recessive disorder, so the authors focused their search on genes with two or more nonsynonymous or splice site variants. Some 42 genes qualified, but 19 of these were known to have multiple copies in the genome. That left 23 genes, and one stood out: ABCG5, an ATP-binding cassette hemitransporter in which the patient had two heterozygous nonsense mutations. A gene, by the way, that had already been linked to the sterol-elimination deficiency (sitosterolemia) that can cause high cholesterol. There was no way the gene could be functional, and targeted sequencing showed that each parent had provided a defunct copy.
New Understanding of the Disease Mechanism
The researchers did some more tests, and the results were consistent with sitosterolemia. So how did they miss it the first time? Well, at the time of diagnosis, 80% of the patient’s diet was breast milk, which is low in plant sterols. At the time of the second test, she was on baby foot (fruits and vegetables), which provided plenty. That explains why the sterol counts were initially low. Why was the cholesterol so high? The authors conclude that in sitosterolemic patients, the severe hypercholesterolemia may reflect an failure to excrete cholesterol into bile, rather than a disruption of cholesterol homeostasis by elevated plant sterols.
Thus, whole-genome sequencing not only provided the correct diagnosis, but may have shed new light on the mechanisms of lipid disease.
References
Rios J, Stein E, Shendure J, Hobbs HH, & Cohen JC (2010). Identification by Whole Genome Resequencing of Gene Defect Responsible for Severe Hypercholesterolemia. Human molecular genetics PMID: 20719861
“At the time of the second test, she was on baby foot ”
Cannibalism?!