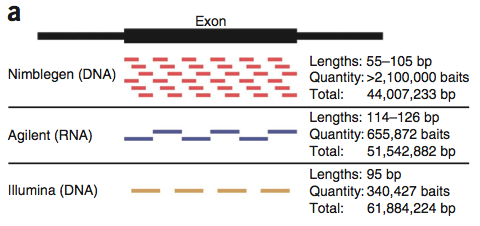
A new study in PNAS from Jay Shendure’s group at the University of Washington describes exome sequencing of 23 prostate cancers. These tumors were derived from aggressive primary tumors or lethal metastases, and propagated in immunocompromised mice as xenografts. For most of the tumors, matched normal DNA was unavailable, so the authors developed a filtering strategy in which the growing catalogs of human sequence variation are employed to identify and remove germline polymorphisms from the lists of tumor genetic variants. Specifically, the authors used pilot project data from the 1,000 Genomes Project, and internally-available variants from ~2,000 additional exomes they’d sequenced. For the majority of tumors, this reduced ~13,500 coding SNVs down to ~350 “nov-SNVs” per tumor (a reduction of 97.4%). The authors readily admit that these nov-SNVs comprise a mixture of:
- Somatic mutations that were present in the original tumor.
- Somatic mutations that occurred during tumor propagation and evolution in the mouse model.
- Germline variants present in the patient’s constitutional genome that are absent from public databases, presumably due to rarity (e.g. private SNPs).
- False-positive variant calls.
Recurrently Altered Gene Filtering
Given a set of mutations from multiple tumors of the same type, the logical next step was to look for genes recurrently altered in the group, since recurrence offers perhaps the best evidence of genes harboring “driver” mutations, which confer advantages for tumor growth and progression, as opposed to “passenger” mutations which do not. The problems for this study were two-fold: First, 16 unique tumors (from unrelated individuals) is a small cohort size with correspondingly small power to identify recurrent alterations. Nothing to be done about that. Second, even looking at just 16 tumors, there were 135 genes harboring non-synonymous nov-SNVs in two or more exomes. A substantial fraction of these are undoubtedly due to rare germline variants missed by the filter, rather than recurrently mutated genes.
To address this, the authors excluded from consideration the 1% of all genes (not just ones mutated in this study) with the highest rate of rare germline variants in control exomes. In other words, they removed genes with the highest rate of germline polymorphism, which I note likely includes (1) genes with high genetic diversity, and (2) genes whose sequence characteristics make them more likely to give rise to false-positive variant calls. The danger of this strategy is that, in principle, genes with high genetic diversity are more prone to mutations, and it’s quite possible that some of these are driver genes for carcinogenesis. Nevertheless, this strategy reduced the list to 104 genes altered in two or more exomes. That’s still too many to tell a story about, so another step was taken.
Using a control set of 1,865 exomes, the authors performed an iterative sampling (I believe this is a bootstrap) to estimate the probability that a given gene would harbor recurrent nov-SNVs that were due to germline variation. Any genes with a germline recurrence probability of 0.001 or higher were excluded from the list, which dropped it sharply down to 20 genes with nov-SNVs in two or more prostate tumors (10 of these were found in three or more).
After all of these steps were taken, the top recurrent gene was TP53, which was altered in 5 of 16 tumors (31.25%). No other gene had as many recurrent hits in the study. This is a vote of confidence for the approach, because TP53 is the one of the most frequently perturbed gene in many solid tumor types, including breast and ovarian cancers. Another believable recurrent gene was GPC6, which encodes a cell surface proteoglycan believed to act as a receptor for growth factors and other signaling molecules. Other recurrent genes highlighted in this study (DLK2 and SDF4) are less convincing. The simple fact is that we don’t know for certain which mutations are truly somatic in the primary tumor, so it’s difficult to draw strong conclusions.
Direct Comparison with Matched Normals
A few of the tumors did have matched normal tissue available, and the authors examined these in detail to assess the accuracy of their germline filtering approach. For three tumors, the authors had (1) mouse xenograft tumor tissue, (2) tumor tissue taken from the patient prior to metastasis, and (3) matched normal tissue. They applied exome sequencing to these to determine set of true somatic mutations (valid mutations) in the original tumor exomes. Valid mutations were compared with the xenograft’s predicted nov-SNVs to determine the number of valid mutations detected (valid detected), the number missed (valid missed), the fraction detected (sensitivity), and the proportion of nov-SNVs that were actually false positives (either germline variants or mis-calls).
| Tumor ID | nov-SNVs | Valid Mutations | Valid Detected | Valid Missed | Sensit- ivity | False Positives |
| LuCap92 | 193 | 56 | 51 | 5 | 91.07% | 73.58% |
| LuCap145.2 | 281 | 122 | 106 | 16 | 86.89% | 62.28% |
| LuCap147* | 2,122 | 2,045 | 1,823 | 222 | 89.14% | 14.09% |
Note that only LuCap 92 was the same tumor tissue that was used to make the xenograft; the other two (LuCap 145.2 and LuCap 147) were neighboring metastases, and presumably closely related to the xenografted tumor. Exome sequencing and germline filtering of the xenograft enabled detection of ~89% of valid somatic mutations across all three cases. This is worrisome, because it means that 11% of valid somatic mutations were removed by the germline filtering strategy. More on that later. Perhaps even more troubling is the inferred false positive rate (fraction of nov-SNVs that are not valid somatic mutations in the tumor), which was ~68% for LuCap 92 and LuCap 145.2.
LuCap 147 is notable in that it was one of three “hypermutated” prostate cancer tumors, with 10-fold the number of nov-SNVs. It also had a lower false-positive rate because there were so many valid somatic mutations to detect. There were no distinctive feature to explain the high number of mutations in hypermutated tumors, though it suggests an acquired defect in DNA repair machinery. As only 15% of tumors had this mutation phenotype, the low false positive rate is an outlier. For most tumors, two thirds of the nov-SNVs obtained by the filtering approach are not valid somatic mutations.
Reasons to Always Sequence the Matched Normal
I have heard it said that sometime in the near future, our catalogs of human genetic variation will be complete enough that we won’t need to sequence matched normal tissue when studying cancer samples. The authors of this study claim that their results give credence to that notion. I respectfully disagree. True, the germline filtering strategy provided a 150-fold enrichment for valid somatic mutations. However, more than half of the final set of nov-SNVs were false positives (not somatic), and 11% of valid somatic mutations were inadvertently removed. I give you, then, my reasons why I believe we should always sequence the matched normal:
- Public databases are not as good as you think. In this study, curated catalogs of sequence variants from known sources (the authors themselves, and the 1,000 Genomes Project) overlapped with 11% of valid somatic mutations, causing their removal. A filter based on the latest dbSNP is even more dangerous because, as some of us have recently discovered, dbSNP contains a lot of somatic (not inherited) mutations. This is because certain cancer projects have submitted their somatic mutation callsets to dbSNP, and these have been accepted. Also, given the low barrier to entry, one should be aware that a lot of dbSNP entries are experimental false positives. Both of these can overlap with mutations in a tumor genome and cause them to be dismissed as germline variants.
- Non-SNV alterations are not amenable to filtering. Tumor genomes acquire insertions, deletions, structural variants, and copy number alterations, some of which may activate oncogenes or disrupt tumor suppressors. Let’s be honest: the databases of non-SNV variants in germline form are woefully incomplete. Unlike SNVs, the coordinates and alleles of larger variants are ambiguous, which makes comparisons to existing variant catalogs very difficult. There are also other types of genetic changes in a tumor, such as loss of heterozygosity (LOH), that will be missed when you don’t know the normal genotype.
- True somatic mutations are exceptionally rare compared to germline variants. Inherited sequence variants occur at a rate of one per 500-1000 base pairs. In contrast, for most tumors, somatic mutations occur at a rate of one per million base pairs. Let’s say you have 20,000 coding variants in a tumor and 98% of those are in dbSNP. That leaves 400 private SNPs that filtering won’t remove, whereas most solid tumors harbor less than 100 somatic coding mutations. In this realistic scenario, only one out of every five post-filtered variants is a somatic mutation.
- Sequencing is cheap, but mistakes are not. Not long ago, you could argue that sequencing matched normals was too costly to be done systematically, even if they were available. That’s no longer the case. A single HiSeq lane gives you enough sequence for two exomes. Why not eliminate the largest source of false-positive mutations – the constitutional genome – by sequencing it as well? It will give you better predictions, and if you go on to validate candidate mutations (as you certainly should), it will probably end up saving you money. Trust me, it’s far better to sequence tumor-normal pairs together, at the same time, same exome platform, ideally same instrument run, to minimize batch effects between them.
Availability of Matched Normals
Of course, sequencing a matched normal sample requires that such material is available. I recognize that this is not always the case. Some of the better-studied cancer cell lines, for example, were made from the tumors of long-dead cancer patients. For less common cancer types, many of the available samples will be frozen or FFPE samples, and getting a matched normal won’t be possible. However, if matched normal tissue is available, I’d argue that it should be assigned for sequencing under identical protocols as the tumor sample. And when you find those germline variants, don’t forget to submit them to dbSNP.
References
Kumar A, White TA, MacKenzie AP, Clegg N, Lee C, Dumpit RF, Coleman I, Ng SB, Salipante SJ, Rieder MJ, Nickerson DA, Corey E, Lange PH, Morrissey C, Vessella RL, Nelson PS, & Shendure J (2011). Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proceedings of the National Academy of Sciences of the United States of America, 108 (41), 17087-92 PMID: 21949389