Next-generation sequencing has immense transformative potential for medicine in the coming decade. Rapid, economical whole-genome sequencing can provide a wealth of information useful for diagnosis, treatment, and even prevention of disease. Very soon (if not already), generating whole-genome sequencing data will be routine. The challenges will lie in accurate variant calling, phasing, annotation, and clinical interpretation.
A new study in PLoS Genetics reports the whole-genome sequencing and detailed genetic risk assessment of a family quartet with a history of familial thrombophilia. There’s a lot to like about this paper, but let me give you the highlights.
- Construction of and alignment to an ethnicity-specific major allele reference sequence yielded improved alignment and more accurate genotyping, especially at disease-associated loci.
- Mendelian inheritance state analysis in the family structure enabled identification and removal of >90% of variants arising from sequencing errors.
- Per-trio phasing, inheritance state of adjacent variants, and population-level linkage disequilibrium data were integrated to provide long-range phased haplotypes.
- By fine-mapping recombination events to sub-kilobase resolution, the authors were able to perform sequence-based human lymphocyte antigen (HLA) typing.
- A curated database of genotype-phenotype correlations made it possible to construct comprehensive genetic risk profiles, including multigenic risk of inherited thrombophilia, common disease susceptibility, and pharmacogenomics.
Advantages of an Ethnically-Concordant Reference Sequence
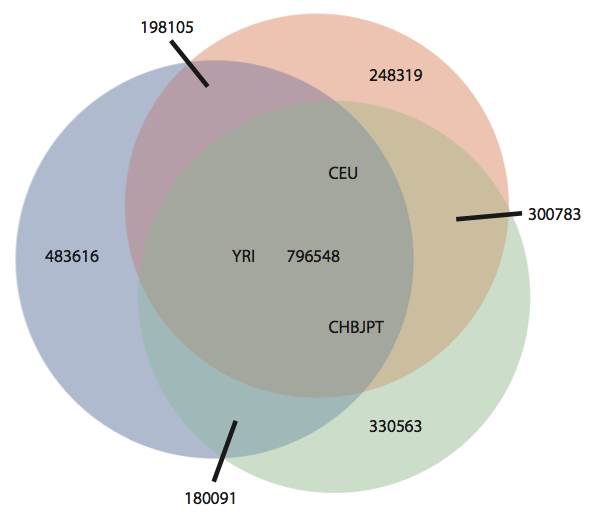
The human reference sequence is a composite, assembled using pooled sequence data from about 20 individuals. Several groups have reported that the current reference harbors a number of biases – some alleles represented are the minority of those present in world populations, and insertions are better represented than deletions. Using SNP genotype data from the 1,000 genomes project (~6-10m loci), the authors of this study developed three ethnicity-specific reference sequences for the CEU (Western Europe), YRI (Sub-saharan Africa), and CEU/JPT (Han Chinese / Tokyo Japanese) populations. They did so by determining the major allele in each population, and swapping it in when the NCBI reference base differed. This resulted in ~1.6 million substitutions for each population reference:
There were almost 800,000 positions where the reference allele was not the major allele in all three populations. Thus, at roughly 10% of SNP positions examined, the NCBI reference sequence contained a minor allele relative to European, African, and Asian populations.
Self-reported ethnicity of the parents in the quartet was northern/western European, a claim largely confirmed by PCA analysis. The authors therefore aligned all genomes to the CEU major allele reference, resulting in a small increase (0.1%) in the fraction of reads mapped by BWA. This seems like a small fraction, but it works out to around 6 million reads across the four samples. Presumably, more reads were mapped because the population-matched reference reduces allele-specific mapping bias (ASMB) against non-reference bases. Next, the authors compared variants to an internally-curated database of genotype-phenotype correlations, identifying 9,389 correlated variants in the family quartet. This number would have been 10,396 if the NCBI reference were used, indicating that 10% of disease-associated markers are in fact major population alleles less likely to contribute to inter-individual variation in disease susceptibility.
The ethnicity-matched reference also enabled a more accurate estimation of population mutation rate (7.8 x 10-4). Using the NCBI reference, this rate was 9.2 x 10-4, indicating that a standard reference sequence yields inflated population mutation rates.
Mendelian Inheritance and Long-Range Haplotyping
Whole-genome sequencing of a “nuclear” family (mother, father, son, daughter) has a number of advantages:
- It enables comprehensive Mendelian inheritance analysis, to facilitate the removal of false-positive variants, isolate putative de novo mutations, and even identify regions of structural variation based on blocks of Mendelian inconsistencies.
- Meiotic crossover sites can be comprehensively surveyed, in this case to sub-kilobase resolution.
- Trio information (each child compared to both parents) helps to phase the variants, in other words, to determine which variants are on the paternal chromosome, and which are on the maternal chromosome. This is especially useful for identifying compound heterozygotes for recessive traits.
- Paired with population linkage information from the HapMap and 1,000 Genomes Project, this information can be used to infer long-range haplotypes. On chromosome 6, the authors used haplotype and population information to accurately determine HLA genotypes for every sample.
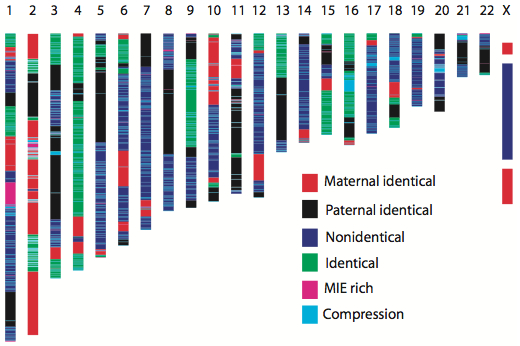
The family information also made possible this fascinating mosaic of chromosomal inheritance:
There are obviously key benefits to having sequence data for everyone in the family. In the future, when clinical sequencing is commonplace, don’t forget to bring your parents along.
Synonymous But Not the Same
One downstream analysis that I particularly enjoyed was that of synonymous coding variants. These variants are often ignored in studies of human genetics, despite a growing body of evidence that they can have translational effects via codon usage bias, mRNA stability, and splice site alteration. The authors developed an algorithm to evaluate these effects for 186 rare, novel synonymous SNPs found in the family. One of these, in the gene ATP6V0A4, is predicted to significantly affect mRNA secondary structure by disrupting a stable “tetraloop” – likely reducing mRNA stability. This is relevant because homozygous loss-of-function variants in this gene have been associated with distal renal tubular acidosis (a disease in which the kidneys don’t remove enough acid into the urine).
Clinical Annotation and Interpretation
The authors build on their previous work to comprehensively annotate clinically-relevant variants in all family members. There’s an extensive amount of work done here, much of it hinging on the authors’ internally-developed, hand-curated database of 16,400 SNPs associated with disease traits. An analysis of rare variants bolstered with evolutionary conservation data highlighted variants in two genes related to thrombophilia: one in the F5 gene, encoding Leiden factor V, with increased risk for thrombophilia, and another in the MTHFR gene (love that gene symbol), which predisposes carriers to hyperhomocysteinemia.
Looking ahead to the probable treatment of family members with blood-thinning medication, the authors next undertook a pharmacogenetic analysis. Perhaps the best-known example of pharmacogenetics is warfarin (coumadin), an oral anticoagulant given to patients at risk for stroke or deep vein thrombosis (DVT). Warfarin was the fifth-most prescribed drug in the U.S. the last time I checked, but it has a narrow therapeutic window. Too little, and it has no anticoagulant effect. Too much, and it can cause internal bleeding. Variants in a number of genes have been associated with warfarin dosing, but two are predominant: CYP2C9, the primary metabolizing enzyme for the drug, and VKORC1, the drug target. In this family, all four members were homozygous for the CYP2C9*1 allele, associated with normal dose, but heterozygous for VKORC1-1639, associated with “therapeutic prolongation” of warfarin response at low doses. Based on these genotypes and patient clinical data, the authors applied the International Warfarin Dosing Algorithm to determine the appropriate dose.
All told, this is an interesting study that clearly involved a substantial amount of work (the pre-print PDF totaled more than 100 pages). Undoubtedly, many of the strategies presented here will be useful as whole-genome sequencing moves into the clinic.
References
Frederick E. Dewey, Rong Chen, Sergio P. Cordero, Kelly E. Ormond, Colleen Caleshu, Konrad J. Karczewski, Michelle Whirl-Carrillo, Matthew T. Wheeler, Joel T. Dudley, Jake K. Byrnes, Omar E. Cornejo, Joshua W. Knowles, Mark Woon, Katrin Sangkuhl, Li Gong,, Madeleine P. Ball, Alexander W. Zaranek, Heidi L. Rehm, George M. Church, John S. West, Carlos D. Bustamante, Michael Snyder, Russ B. Altman, Teri E. Klein, Atul J. Butte, & Euan A. Ashley (2011). Phased whole genome genetic risk in a family quartet using a major allele reference sequence PLoS Genetics, 7 (9)
Thanks. With regards just this claim:
it is not clear from Figure 4C that the HLA types are accurate –
[a] the types are mostly homozygote at each gene, which is extremely unlikely
[b] (perhaps as a result) they appear to misinherit wildly.