Two studies in the journal Science demonstrated that genes in the hypoxia-inducible factor (HIF) oxygen signaling pathway have undergone strong, recent positive selection in Tibetan highlanders. One study was a genome-wide scan using SNP arrays; the other a large-scale exome sequencing effort. The exome study was particularly interesting; using the Nimblegen 2.1M exon capture array and Illumina GAIIx instruments, Yi et al sequenced the exons of nearly 20,000 genes (92% of CCDS) in 50 unrelated Tibetans.
Exome Sequencing Summary
To my knowledge, this represents the largest published study of human exome sequencing to date. The main text in the report to Science was necessarily brief, so I used the supplemental materials to glean the following information:
|
The production numbers are consistent with a single lane of 2×75 bp reads (3.4 Gbp) per exome. The low mapping rate (68%) is slightly alarming, but I’d guess (hope) that only uniquely mapped reads are counted here. The on-target mapping rate, a measure of capture specificity, was 68%, well within the expected enrichment of large-scale capture technologies.
Highly Variable Coverage Across Samples
I do feel obligated to point out that while the average target depth was 18x, which seems appropriate for variant calling, the actual target depth varies widely across the 50 samples. Here’s my plot of target coverage breadth (% of bases) by average target depth (redundancy) using data from supplemental table 1:
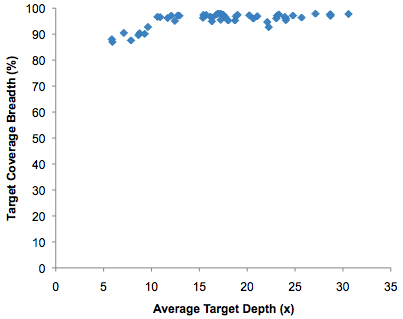
Almost every sample reaches 90% coverage breadth, but 7 of them have less than 10x coverage on average. This will undoubtedly affect the ability to call variants accurately, though only a statistician might be able to extrapolate the effects of such variable coverage on the study’s outcome.
Searching for Selection
To look for evidence of positive selection for altitude, they compared SNP allele frequencies to between Tibetans and 40 Han Chinese whose genomes were sequenced to low (4x) coverage as part of the 1,000 Genomes Project. About 100,000 high-confidence SNPs (>99% probability) were called in the Tibetan samples. A subset (53/56) were validated by Sanger sequencing, suggesting that ~95% of sites are valid polymorphisms. Allele frequency estimates showed an excess of low-frequency variants, particularly among nonsynonymous SNPs.
Using synonymous sites in both populations, the population historical modeling estimated that Tibetans and Han Chinese diverged 2,750 years ago, with Han expanding from a small initial population, and Tibetans shrinking from a larger ones. Migrational evidence suggests that Han Chinese migrated from the Tibetan region, with recent admixture in the opposite direction.
Exon Targets, Intron Findings
Intriguingly, though the “exome” sequencing strategy focused on coding regions, no amino-acid changing variants differed by more than 6% between Han and Tibetan populations. Fortunately, hybrid selection (capture) also captures some of the noncoding regions that flank target exons. This happens because randomly sheared DNA fragments (200-250 bp) may overlap both exon and intron sequence, yet still have enough sequence overlapping a probe to be captured. This creates a “shoulder” of coverage upstream and downstream of target exons, often in intronic or UTR sequences.
This side-benefit of exome capture proved serendipitous because intronic sequences harbored the most divergent SNP between Han (9% frequency) and Tibetan (87% frequency) populations. The gene in question was endothelial PAS-domain protein 1 (EPAS1), also known as hypoxia-inducible factor 2-alpha (HIF2A). Hypoxia in the name of a candidate gene for high altitude adaptation was a good sign. A protein-stabilizing mutation in EPAS1 had already been linked to erythrocytosis, suggesting a possible link between this gene and red blood cell production.
Even more promising was the fact that another study published in the same issue of Science had pinpointed the same gene by high-density SNP array genotyping. The irony here is priceless: an expensive exome sequencing project finds an intronic SNP, implicating a gene that was just as easily identified by genotyping. Of course, if the relevant haplotypes had been comprised of rare variants – ones absent from the Han population and not covered by current SNP arrays – only one group would have identified this gene, and the other would have gone home empty-handed.
Perspective
Storz, J. (2010). Genes for High Altitudes Science, 329 (5987), 40-41 DOI: 10.1126/science.1192481
Reports
Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, Prchal JT, & Ge R (2010). Genetic evidence for high-altitude adaptation in Tibet. Science, 329 (5987), 72-5 PMID: 20466884
Yi X, Liang Y, Huerta-Sanchez E, et al. (2010). Sequencing of 50 human exomes reveals adaptation to high altitude. Science, 329 (5987), 75-8 PMID: 20595611
Speaking of exomes and positive selection, this paper by Josh Akey was published in GR http://bit.ly/9DZUbo
Excellent synopsis. Thanks!