Exome sequencing, which targets the exons of known protein-coding genes in the human genome, is being widely employed by the research community to determine the genetic basis of inherited disease. It’s rapidly becoming the frontline research tool in GWAS follow-ups, tumor-normal comparisons, and studies of Mendelian disorders.
 The authors of some publications have mistakenly referred to this as “whole” exome sequencing, which is a bit of a misnomer.
The authors of some publications have mistakenly referred to this as “whole” exome sequencing, which is a bit of a misnomer.
Although the vast majority of protein-coding bases in the human genomes are represented on current exome sequencing kits, it’s certainly not the whole exome or even the whole known exome. Some genes are poorly covered; others are missing entirely.
Exome Sequencing’s Dual Effect
The widespread application of exome sequencing to a vast number of samples and cohorts has two complementary effects. First, as we’ve already seen, it has enabled the rapid identification of many disease genes. For the most part, these are genes in which deleterious genetic variants confer highly penetrant susceptibility to a rare inherited disorder.
The second effect of widespread exome sequencing, one that many of us saw coming, is now beginning to emerge: exome sequencing is not enough. Negative results, such as the failure to uncover the cause of a Mendelian disorder using traditional exome sequencing and analysis, often go unpublished. It’s difficult to get such results into a high-impact journal, and most researchers would rather wait until they can explain the phenotype (usually by whole-genome sequencing or a non-traditional analysis).
Even if I didn’t have first-hand experience of exome failure, there are plenty of subtle signs out there.
- The number of Mendelian disorders solved is far less than the number we know have been studied, especially at the NHGRI-funded Mendelian centers.
- Manufacturers of exome reagents are engaged in an arms race to include more and more targets, now including many noncoding sequences such as UTRs.
- A growing number of researchers are employing whole-genome sequencing, even though the cost hasn’t changed dramatically in a couple of years.
Why Exome Sequencing Studies Fail
So the evidence suggests that the exome strategy is often a let-down. This is a bit surprising for many of the rare, highly penetrant Mendelian disorders where sufficient samples from large pedigrees are available. In a previous post, I outlined 6 possible explanations for elusive Mendelian disease genes. One of those, in the hard-to-tackle category, is the very plausible possibility that a noncoding functional variant is responsible.
Tiering the Genome
In AML1 and subsequent studies, we devised a relatively simple system for classifying mutations in a cancer genome based on annotation information:
- Tier 1 mutations affect known protein-coding exons and noncoding RNA genes.
- Tier 2 mutations map to conserved regions and regulatory sequences that aren’t coding but might have regulatory potential.
- Tier 3 mutations map to other noncoding but unique regions of the genome
- Tier 4 mutations are in repetitive, non-unique regions that we tend to ignore.
It’s not a perfect system, but it’s simple and defensible: tier 1 mutations are most likely to affect protein structure and/or function, and also the easiest to interpret, so they’re the top priority. After that, tier 2 seems to be the next obvious place to look for causal mutations. Due to rapidly evolving technologies and a growing body of knowledge about the genome, the definition of such tiers is quite dynamic.
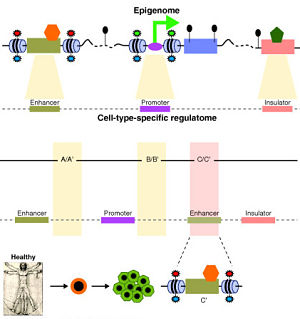
Regulatory Sequences in the Genome
High-throughput technologies — including microarrays, next-gen sequencing, and others — have proven powerful tools to characterize the regulatory sequences in the genome. Many groups such as the ENCODE consortium are characterizing human genome content and function using technologies such as:
- RNA-Seq, to measure gene and small RNA expression in different tissues or at different time points
- CHiP-Seq, to identify genomic regions bound by histones, transcription factors, and other specific proteins.
- DNAse sequencing and footprinting, to characterize regulatory regions based on sensitivity to or protection from cleavage by DNase I enzymes.
- Bisulfite sequencing, to measure the pattern and extent of DNA methylation across the genome.
- Chromosome conformation capture (3C) and sequencing, to find long-range interactions enabled by DNA’s three-dimensional structure.
These types of experiments generate a wealth of data about regulatory activity in genomes. While studying each of these independently is certainly informative, integrative analysis will be required to elucidate how all of these different regulatory mechanisms work together. That will require robust statistical models, substantial computing resources, and productive collaboration among research groups. Provided that we, the research community, can bring all of these things together, what should ultimately emerge will be a far more complete understanding of how the genome works.
That is ultimately what will be required before we can identify and implicate noncoding functional variants underlying genetic diseases.
References
Rada-Iglesias A, & Wysocka J (2011). Epigenomics of human embryonic stem cells and induced pluripotent stem cells: insights into pluripotency and implications for disease. Genome medicine, 3 (6) PMID: 21658297
Of course, there is the small issue that all of the other methods described above require a specific tissue sample. For some tissues such as brain, for example, this will rarely ever be practical in a clinical setting. For other tissues, it will be interesting to see if we eventually find these methods diagnostically relevant enough to warrant doing biopsies, etc., rather than just being able to do them if such samples already exist.
Paul, a very good point. Thanks for bringing it up!