 Upon completion of the draft sequence of the human genome, it quickly became apparent that only a fraction of DNA (1-3%) comprised known protein-coding genes. This gave rise to the somewhat mistaken notion that ~98% of the genome contains “junk DNA” with no purpose or function. We know, of course, that noncoding portions of the human genome contain functional elements contributing to phenotypic variation. We simply didn’t know what they were.
Upon completion of the draft sequence of the human genome, it quickly became apparent that only a fraction of DNA (1-3%) comprised known protein-coding genes. This gave rise to the somewhat mistaken notion that ~98% of the genome contains “junk DNA” with no purpose or function. We know, of course, that noncoding portions of the human genome contain functional elements contributing to phenotypic variation. We simply didn’t know what they were.
In 2003, two large-scale consortia undertook ambitious efforts to understand the variation and function in the human genome. The first of these efforts took the form of the International HapMap Project and later the 1,000 Genomes Project, whose goal was to characterize human genetic variation. A second, perhaps less-hyped initiative (called the Encyclopedia of DNA Elements, or ENCODE) set out to delineate all functional elements in the human genome.
ENCODE Papers Are Out
This month, the tremendous efforts of the ENCODE Project Consortium have borne fruit with 30 publications in Nature, Genome Research, and Genome Biology. The main paper in Nature describes the initial production and analysis of 1,640 datasets, encompassing diverse experiments within several cell types to characterize regions of RNA transcription, protein-coding sequences, transcription factor binding sites, chromatin structure, and DNA methylation regions in the human genome.
The ENCODE Dataset
Here’s a brief summary of the dataset described in the main paper:
| Element | Approach | Description |
| Genes and RNAs | Annotation | Automated and manual annotation to produce the GENCODE reference gene set (v7) of protein-coding genes, pseudogenes, and non-coding RNAs. |
| Transcribed regions | RNA-seq | An extensive RNA expression catalogue from different cell lines and multiple subcellular fractions. |
| Transcription start sites | CAGE-seq | 5′ cap-targeted RNA isolation and sequencing identified 62,403 high confidence TSSs. |
| Full-length RNAs | RNA-PET | Simultaneous capture of RNAs with both a 5′ methyl cap and a poly-A tail |
| Protein-coding regions | Mass spec | Peptide sequences identified by mass spectrometry in two cell lines (K562 and GM12878) |
| TF binding sites | CHiP-Seq | Mapping binding locations of 119 transcription factors and several RNA polymerase components in 72 cell types. |
| Open chromatin | DNase-Seq & footprinting, FAIRE | Sequencing of DNase I hypersensitive sites in (125 cell types), FAIRE analysis of reduced nucleosome crosslinking (25 cell types), and genomic DNase I footprinting (41 cell types). |
| Histone modification | CHiP-Seq | Assays for up to 12 histone modifications and variants in 46 cell types |
| DNA methylation | RRBS | Reduced representation bisulfite sequencing to profile 1.2m CpGs in 82 cell lines and tissues |
| Chromosome interactions | 5C and ChiA_PET | Chromosome conformation capture carbon-copy (5C) in a targeted 1% (4 cell types). Genome-wide chromatin interaction analysis with paired tag sequencing (ChIA-PET) of PolII CHiP in 5 cell types. |
Study Highlights
It would take me weeks to carefully review and digest these 30 publications. There’s just so much discovery that was enabled by the massive dataset. Here, I’ll give you some highlights from the main paper. In the next week, as time allows, I’ll follow up on some of the companion studies that I found particularly interesting. Here we go.
GENCODE Annotation
Gencode version 7 is the default annotation for ENCODE integrated analysis. It includes:
- 20,687 protein-coding genes with 6.3 alternatively spliced transcripts per locus, whose coding exons encompass 1.22% of the genome.
- 11,224 pseudogenes, of which 863 were transcribed and associated with active chromatin.
- 8,801 small RNAs and 9,640 curated long ncRNAs
Transcribed and Protein-coding Regions
One of the most striking findings of the ENCODE pilot project was that the genome is pervasively transcribed; an estimated 60-80% of bases were present in at least one transcript. That observation has been confirmed and extended using the latest in RNA sequencing technologies:
- 62% of genomic bases are reproducibly represented in sequenced long RNA molecules or GENCODE exons
- CAGE-seq revealed 62,403 transcription start sites (TSSs) of which 27,362 (44%) were within 100 bp of the 5′ end of a GENCODE transcript or known mRNA transcript.
- A significant portion of coding and non-coding transcripts were processed into stable small RNA precursors — tRNA, miRNA, snRNA, and snoRNA — and their processed products tend to align with the capped 5′ ends of long transcripts.
I think that this latter point may suggest that small RNAs are generally processed from larger transcripts and, once processed, their 5′ ends line up right where 5′ capping occurs. If I understand this correctly, it means that the final product is a capped (and therefore stable) small RNA.
Protein-binding Regions
ChIP-Seq is a powerful tool to study genetic regulation, because it enables one to accurately identify regions of DNA bound by specific proteins, such as transcription factors or RNA Polymerase II components. The authors applied CHiP-Seq with 119 different DNA-binding proteins (including 87 sequence-specific transcription factors) to 72 cell types, finding:
- 636,336 binding regions spanning 231 megabases (8.1% of the genome) were enriched for DNA-binding across all cell types
- 86% of DNA segments occupied by sequence-specific transcription factors contained a strong DNA binding motif; in most cases (55%) the known motif for the transcription factor was the most enriched.
- CHiP-Seq peaks lacking cognate binding motifs had 21% lower median scores, and most (82% of them) had high-affinity recognition sequences for other factors.
This last observation indicates that CHiP-Seq peaks that didn’t contain known binding motifs are either lower-affinity sites or were captured through the transcription factor’s interaction with other proteins.
Open Chromatin and DNAase I Hypersensitivity
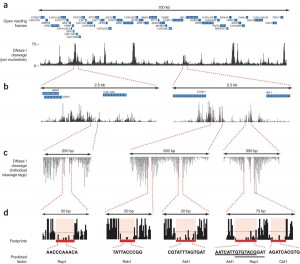
Regulatory elements tend to occur in regions of open chromatin (euchromatin), where they are accessible to transcription factors and other proteins. In contrast, closed chromatin (heterochromatin) is inaccessible because it’s tightly wrapped around nucleosomes. When chromatin preparations are exposed to formaldehyde, those nucleosomes are physically cross-linked to the heterochromatic DNA wrapped around them. A phenol extraction then allows one to sequence and map the crosslinked DNA fragments. It then becomes possible to search for the reduced nucleosomal crosslinking associated with open chromatin and regulatory DNA. This technique, called formaldehyde-assisted isolation of regulatory elements (FAIRE), enabled ENCODE to map 4.8 million sites of reduced crosslinking across 25 cell types.
The DNase I restriction enzyme cleaves regions of open chromatin, and it preferentially does so at places where, nearby, there are non-histone proteins bound to DNA. Sequencing the cut-points of DNase I (DNase-Seq) identifies these DNAse I hypersensitive sites (DHSs), which are about 200 bp long. Interestingly, within each DHS are short (10 bp) sequences that were not cleaved, because they were protected by the bound protein. This process, called genomic DNase I footprinting, makes for a clever system of isolating isolating cis-regulatory sequences, and is nicely illustrated in this figure:
Employing DNase-Seq and genomic DNase I footprinting, ENCODE found:
- 2.89 million unique, non-overlapping DHSs in 125 cell types. Many coincided with the sites of reduced nucleosome crosslinking identified by FAIRE.
- 8.4 million distinct DNase I footprints across 41 cell types. Motif discovery in these recovered 90% of known transcription factor motifs, plus hundreds of novel (and evolutionarily conserved) motifs.
Many of the novel motifs displayed occupancy patterns that were highly cell-type-specific, similar to the patterns observed among major developmental and tissue-specific regulators.
Epigenetic Codes: DNA Methylation and Histone Modification
Methylation of cytosine bases is an epigenetic mechanism by which gene expression is regulated. Methylation of promoters (which are often rich in CpG dinucleotides) is usually associated with repression, whereas methylation of genic regions typically correlates with transcription. ENCODE used reduced representation bisulfite sequencing (RRBS) to profile DNA methylation at 1.2 million CpGs in each of 82 cell lines, finding that:
- 96% of CpGs exhibited differential methylation in at leas one cell or tissue type
- Levels of DNA methylation correlated with chromatin accessiblity
- The CpGs with the most variable methylation levels were more often in gene bodies and intergenic sequences than in upstream regulatory regions.
This last observation seems to indicate that promoter methylation does not play a significant role in regulation of gene expression between tissues.
Regions of Chromosome Interaction
We often think of the human chromosomes as linear, anaphase-type bodies, a notion reinforced by the graphical depiction of chromosomes as linear two-dimensional objects on, say, the UCSC genome browser. In truth, human chromosomes are tightly organized and packed into the nucleus, a situation which causes physical interaction between regions that might be hundreds of kilobases apart. This interaction is thought to be important in regulation of gene expression. To study it, ENCODE utilized two approaches based on chromosome confirmation capture (3C), in which physically interacting regions are cross-linked by formaldehyde, and isolated from non-interacting chromatin with restriction enzymes. Isolation and sequencing of the cross-linked regions (by carbon-copy 3C, or “5C”), enables characterization of interactions with base-level resolution. A complementary approach, chromatin interaction analysis with paired-end tag sequencing (wonderfully abbreviated ChIA-PET), lets you first enrich for chromatin bound by a specific protein. In ENCODE’s case, they performed ChIA-PET of PolII-enriched chromatin in five cell types.
- By applying 5C to a targeted 1% of the genome in 4 cell types, ENCODE discovered hundreds of significant interactions
- On average, 3.9 distal elements interact with a TSS, and 2.5 TSSs are associated with each distal element
- In K562 cells, there were 127,417 promoter-centered chromatin interactions, 98% of which were intra-chromosomal
- The majority of involved promoters participated in multi-gene interaction complexes (often spanning megabases) including promoter-promoter and promoter-enhancer interactions.
As the authors put it, these analyses “portray a complex landscape of long-range gene-element connectivity across ranges of hundreds of kilobases to several megabases, including interactions among unrelated genes.” As if we needed more genetic complexity!
Conclusion: A Note on Big Science
Big science has yielded some of the most important advances in human genetics and genomics. Efforts such as the HapMap Project and 1,000 Genomes Project have characterized human genetic variation at an unprecedented scale. The Cancer Genome Atlas is building a road map of the genetic and molecular properties of common cancers. And now, the ENCODE Consortium has provided us exactly what they promised: an encyclopedia of the functional elements in the human genome.
References
ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489 (7414), 57-74 PMID: 22955616
Dear Dan, great work and greatthanks for it. It is really not an easy task to pass throughall these 30 papers. And you substabtially facilitate this job. Thank you once more.
Maria Obolenskaya
Maria, I’m glad I could help. Thanks for reading!