Now online at Nature is the most comprehensive molecular portrait of human breast tumors published to date. The Cancer Genome Atlas study encompasses more than 500 primary tumors representing the four intrinsic subtypes of breast cancer: Luminal A, Luminal B, Her2-enriched, and Basal-like. Using a suite of high-throughput platforms, TCGA characterized various aspects of these tumors at the molecular level:
- Somatic mutations and germline susceptibility variants (exome sequencing)
- Genomic copy number (SNP and CGH arrays)
- DNA methylation
- microRNA expression (sequencing)
- Gene expression (mRNA arrays)
- Protein expression (reverse-phase protein arrays)
Many of the key insights in this study were enabled through integrative analysis across these platforms. The findings suggest a highly heterogeneous disease whose intrinsic subtype was apparent not only by gene expression, but by mutational and DNA copy number profiles as well. Only three genes (TP53, PIK3CA, and GATA3) were mutated in 10% or more of all tumors; grouping them by gene expression (intrinsic) subtype uncovered a number of subtype-specific events. New recurrently mutated genes, two novel protein expression subtypes, and a previously unrecognized connection to ovarian cancer are just a few of the tantalizing results. Let’s get started.
Primary Breast Tumors
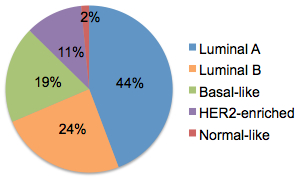
There were 507 primary tumors in the main analysis set; these were the focus of most analyses in this study. The breakdown by intrinsic [gene expression] subtype is generally representative of the disease incidence as a whole:
Luminal A tumors are the most prevalent (44% in this study), followed by luminal B (24%). Both of these subtypes are estrogen receptor (ER) positive, with luminal A carrying the most favorable prognosis. Basal-like tumors compose 19% of the set; these overlap considerably with so-called triple negative breast cancers, which are clinically negative for estrogen receptor (ER), progesterone receptor (PR), and Her2 receptor (HER2). Another 11% are of the Her2-enriched subtype, a group whose prognosis has improved since the development of targeted therapies against Her2 (e.g. Herceptin). Finally, 2% of tumors were of the rare normal-like subtype, which couldn’t be comprehensively studied with so few samples.
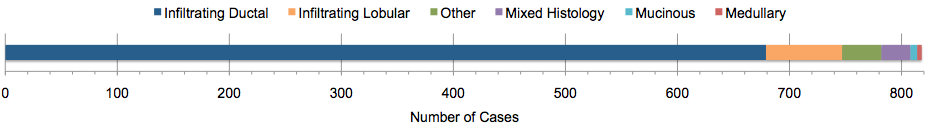
Another way to look at breast cancer tumor is by histology type, which reflects the cell of origin:
Most often, breast cancer arises from the mammary ducts (ductal) or milk-producing lobules (lobular), though other less common histology types were represented as well.
Somatic Mutations in Breast Cancer
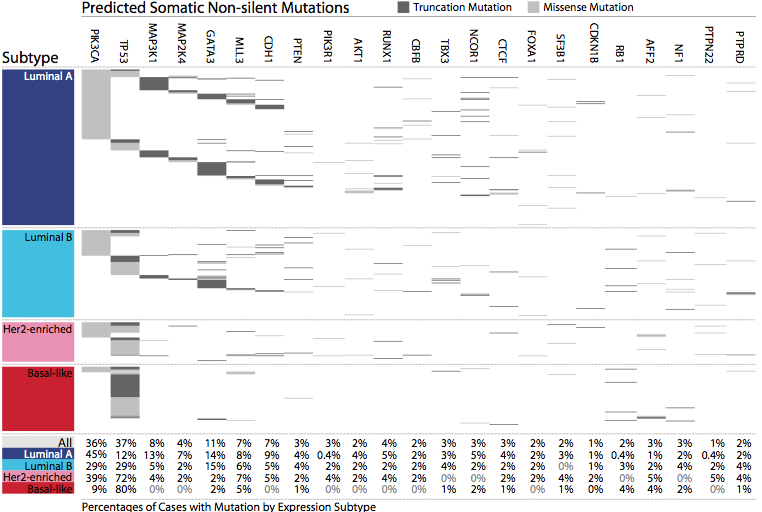
Figure 1 nicely encapsulates the characterization of breast cancer genomes as a whole and by intrinsic subtype.
The most frequently mutated gene overall was TP53 (37% of cases), followed closely by PIK3CA (36%). Mutually exclusive mutations in MAP3K1, MAP2K4, and GATA3 were prevalent in luminal A and luminal B subtypes, but almost absent from the others. In contrast, TP53 mutations were frequently observed in Her2 (72%) and Basal-like (80%) subtypes. Mutations in E-cadherin (CDH1) were frequent across subtypes but correlated almost perfectly with lobular tumors.
The observed mutation rate in coding regions seems to reflect the aggressiveness of breast cancer subtypes: lower in luminal A (0.84 mutations/Mbp) than luminal B (1.36 mutations/Mbp), higher in Her2-enriched (2.05 mutations/Mbp) and Basal-like (1.68). The number and extent of copy number alterations, too, followed this trend.
Analysis with the Mutation Significance In Cancer (MuSiC) package identified 35 significantly mutated genes (SMGs). Among these were nearly all genes previously implicated in breast cancer (PIK3CA, PTEN, AKT1, TP53, GATA3, CDH1, RB1, MLL3, MAP3K1 and CDKN1B) and a number of novel SMGs, including:
- TBX3, which is mutated in ulnar-mammary syndrome and involved in mammary gland development
- Ubiquitous transcription factors CTCF and FOXA1
- RUNX1 and CBFB, both rearranged in acute myeloid leukaemia and interfering with haematopoietic differentiation
- PIK3R1, in which mutations clustered in the PIK3CA interaction domain (similar to glioma and endometrial cancer).
- Splicing factor SF3B1, previously reported in myelodysplastic syndromes and CLL
- Protein tyrosine phosphatases PTPN22 and PTPRD
A statistically significant exclusion pattern among PIK3R1, PIK3CA, PTEN and AKT1 mutations (P = 0.025) reflects the well-known activation of PI3K signaling in breast cancer.
Basal-Ovarian Connection
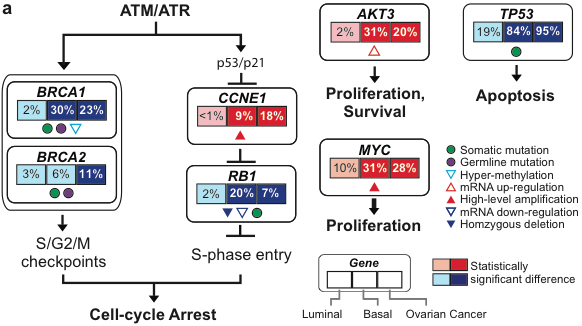
One of the most intriguing findings comes from a comparison of breast cancer subtypes to other common cancers. Figure 5 shows how key pathways are altered in luminal (first % box) and basal (middle box) breast cancer and ovarian cancer (right box).
Among these and other results integrating multiple platforms, it’s apparent that Basal subtype tumors more closely resemble ovarian tumors than other breast cancer subtypes. This raises the exciting possibility that therapies for ovarian cancer might benefit patients with Basal breast tumors, and vice-versa.
I won’t spoil any more of this outstanding paper, but recommend that you read it yourself online at Nature where it’s open access.
References
The Cancer Genome Atlas Network (2012). Comprehensive Molecular Portraits of Human Breast Tumors Nature DOI: 10.1038/nature11412

It was an interesting article. The main focus seems to be more on mutation and other genetic abnormalities that lead to cancer. I am wondering how epigenetics figures into the equation. There appears to be a lot of work in this area with relation to cancer.
Thanks