Triple-negative breast cancer (TNBC), a tumor type defined by its lack of estrogen receptor, progesterone receptor, and Her2 (ERBB2) amplification, accounts for 16% of breast cancers. This clinically defined tumor type overlaps substantially but not completely with “basal-like” breast cancer, a classification based upon gene expression signature. This is a highly heterogeneous disease with a higher risk of recurrence in the absence of systemic therapy.
This month in Nature, researchers from BC Cancer Agency have characterized the landscape of genomic aberrations in 104 TNBC cases with a combination of whole-genome sequencing, exome sequencing, RNA-seq, and high-density SNP arrays. Using ultra-deep targeted resequencing, the authors validated ~2,500 somatic mutations and characterized their frequencies among heterogeneous clonal populations in each tumor.
This is a complex study that’s hard to digest (the supplemental material had over 140 pages – come on, is that really necessary?) so I’ll do my best to break it down. I believe there are three highlights: frequent gene alterations in TNBC, under-representation of mutations in mRNA sequences, and a continuous distribution of mutation frequencies within tumors.
Genetic Alterations in Triple-Negative Breast Cancer
The most frequently mutated gene should be familiar to you: TP53, which harbored validated somatic mutations in 62% of basal and 43% of non-basal TNBCs. Other patterns of alteration were as follows:
- Significantly mutated genes included TP53, PIK3CA, RB1, PTEN, MYO3A, and GH1. Here, significant means that the gene harbored more mutations than expected from background random mutation processes. The larger the gene, the more likely it is to catch a random mutation. That’s why USH2A, which was mutated in 9.2% of cases, was not significant (it’s a large gene).
- Recurrent but not statistically significant mutations were observed in the synuclein genes (SYNE1/SYNE2), BRCA2, BRAF, NRAS, ERBB2, and ERBB3.
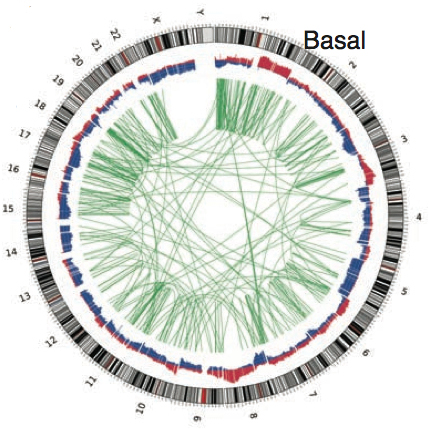
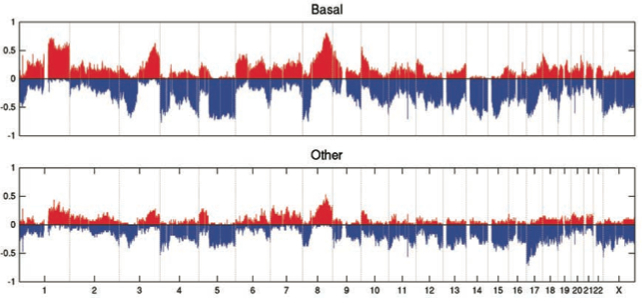
Somatic Copy Number Alterations (CNAs)
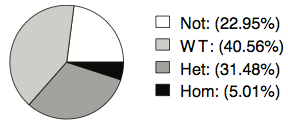
Expression of Somatic Mutations
- 40.56% of genes where only the wild-type allele is expressed. Here, it’s possible that the mutation alters mRNA expression or stability and thus only the non-mutated allele is seen.
- 31.48% where both alleles are expressed. The mutation may not affect expression, but it could still alter the translation or function of the encoded protein.
- 5% where only the mutant allele is expressed. This could be due to genomic loss of the wild-type allele (LOH), mutations on the X-chromosome (one copy of which is inactivated), or even a gain-of-function mutation causing aberrant gene expression.
Continuous Distribution of Somatic Mutations
With ultra-deep targeted sequencing, it’s possible to estimate the allele frequency of a somatic mutation with high accuracy, and from that, to infer the relative proportion of tumor cells harboring that mutation. A heterozygous founder mutation, for example, would be present in virtually all tumor cells and have a mutation frequency of 50% in diploid cells. Perhaps surprisingly, the authors find that somatic mutations occur at a continuous distribution in TNBC, and this appears independent of copy number alterations and tumor cellularity.
Part of this observation may technical in nature (i.e. false negatives in mutation discovery). However, this phenomenon has been noted in other epithelial cancers suggesting that the mutation content of cells within a single tumor may be differently shaped by biological processes and mutational mechanisms. It reinforces the notion that tumors (and TNBC in particular) are not a homogeneous mass of identical cells, but a collection of distinct sub-populations of cells evolving somewhat independently of one another. This is probably why they’re sometimes difficult to eliminate: you might destroy most of the subpopulations with therapy, but one or more minor clones could persist.
References
Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, & Aparicio S (2012). The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature PMID: 22495314