I’ve returned from Baylor’s Human Genome Sequencing Center (HGSC), where earlier this week colleagues from Baylor, Boston College, the Broad Institute, University of Michigan, NCBI, and EBI converged for a face-to-face meeting on Pilot 3 of the 1,000 Genomes Project. Unlike pilots 1 and 2, which emphasized whole genome sequencing to low or high coverage, respectively, in Pilot 3, the exons of 1,000 genes (~1.5 Mbp total) were selectively targeted for sequencing by capture technologies.
Capture, Exons, and the Exome
For the genome centers, this pilot was one of the first applications of relatively new technologies to enrich for particular regions of the genome. The idea is that by focusing on the exons of protein-coding genes, one can maximize the return of sequencing because variation in those regions is [presumably] more likely to be phenotypically relevant. A post by fellow blogger Keith Robison of Omics! Omics! discusses how capture technologies have recently scaled to offer “exome sequencing” and wonders if this approach will miss important non-coding variation.
While the question of which genomic regions harbor phenotypically-relevant variation is a subject of open debate, I think that Pilot 3’s focus is more technological than biological. It motivated Baylor, Broad, WashU, and Sanger centers to push developing capture technologies into production. Perhaps the most important aspect of this project, as Carrie Sougnez of the Broad Institute put it, is that Pilot 3 “helped us learn how to do capture.”
Cross-Platform and Cross-Pipeline Comparisons
In the face-to-face meeting, Kiran Garimella of the Broad Institute and Gabor Marth of Boston College presented some comparisons of variant calls across platforms and across BAM-generation pipelines. The results, surprisingly, were similar across most of the approaches in terms of the variants that were detected. Comparisons of BAM files generated by different pipelines (Broad’s and Baylor’s, for example) revealed few differences. One exception, however, was the aggressive marking of PCR duplicates in 454 data by Baylor’s MarkDuplicates algorithm, which reduced the number of [false-positive] SNP calls. Matthew Bainbridge of Baylor has already been generous enough to share this algorithm with other centers.
Overall, the Pilot 3 variant calls are looking good – dbSNP concordances in the 70-80% range or higher, and transition/transversion ratios of about 3-3.50 – and consistent across 454 and Solexa data from multiple centers.
Validation and Biological Significance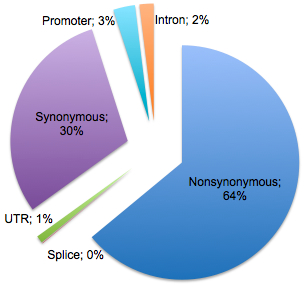
As with any SNP discovery project, validation is a key step, and the decisions of how to validate thousands of variants across hundreds of samples are non-trivial. Much of the face-to-face meeting discussions were devoted to coming up with a validation plan. While we don’t yet know for certain how many of the ~80,000 novel putative variants discovered in Pilot 3 are real, the results look promising. As expected, novel variants tend to be rare – found in just one or a few individuals in the study. Yet our strategy of capture-based sequencing to target exons seems to be paying off, because more than half of the novel variants are predicted to alter protein sequence (nonsynonymous) or mRNA splicing.
Although there’s a lot of work yet to be done, it’s clear to me that this Pilot, and the 1,000 Genomes Project as a whole, will yield a tremendous wealth of new knowledge about sequence variation in the human genome.
The transition/transversion ratio seems high.