Next-generation sequencing promises to reach unprecedented levels of throughput this year, driving down the cost of sequencing dramatically. Somewhere between the GridION, the Ion Proton, and the HiSeq2500, we may see the first single-day, $1,000-per-genome technologies in 2012. Even so, a 90% reduction in sequencing cost this year will not magically solve all medical problems, even the ones that are clearly genetic. We are already reaching a point where getting enough sequencing coverage and finding the variants no longer present a significant problem. Instead, the field of human genetics faces three significant challenges as we enter an era of ultra-low-cost sequencing.
1. Obtaining Sufficient, Relevant, Consented Samples
2. Clinical Annotation of Genetic Variants
3. Interpretation of Complex Genomes
Obtaining Sufficient, Relevant, Consented Samples
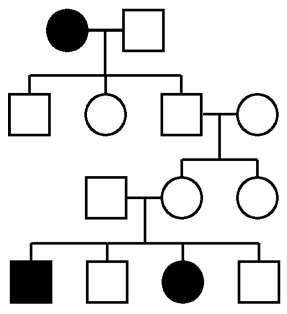 Samples will become a major challenge. Specifically, obtaining sufficient numbers of high-quality, accurately phenotyped, properly consented samples for sequencing. I know for a fact that many, many studies are not facing a bottleneck at sequencing capacity but at sample collection, consent, and banking. There are even internationally renowned cancer centers where banking tumor samples and patient blood samples is not a standard or required practice for oncologists. A sad reality is that, every day, people succumb to diseases such as cancer, metabolic syndromes, and heart disease where genetics undoubtedly plays a role. Those samples, if not banked, are lost to the world of science.
Samples will become a major challenge. Specifically, obtaining sufficient numbers of high-quality, accurately phenotyped, properly consented samples for sequencing. I know for a fact that many, many studies are not facing a bottleneck at sequencing capacity but at sample collection, consent, and banking. There are even internationally renowned cancer centers where banking tumor samples and patient blood samples is not a standard or required practice for oncologists. A sad reality is that, every day, people succumb to diseases such as cancer, metabolic syndromes, and heart disease where genetics undoubtedly plays a role. Those samples, if not banked, are lost to the world of science.
The good news is that there are many excellent cohorts out there. There are entire populations that have been catalogued, sampled, and followed-up-with over the course of decades, with huge amounts of qualitative and quantitative clinical data. The commoditization of sequencing means that the proprietors of these cohorts will have their choice of sequencing providers. Informative samples, especially those from patients suffering from rare inherited disorders, will be in high demand. Tumor samples will be fought over by researchers, drug companies, and the treating physicians. In a world of cheap sequencing, samples are the new commodity.
Clinical Annotation of Genetic Variation
With long enough reads and sufficient coverage, finding mutations will no longer be a problem. The new challenge will be in assessing their functional significance and determine which have clinical relevance.
Imagine a breast cancer patient whose germline and tumor genomes have been sequenced to high depth. You have the full spectrum of germline/somatic mutations, copy number alterations, and structural variants. And you also have so many questions:
- • Which of these mutations are drivers? Which are passengers?
- • What do the variants say about diagnosis or prognosis?
- • Are there any clinically actionable mutations?
- • Have any been seen before in this tumor type, or other tumor types?
- • Are there germline susceptibility variants that predisposed this patient to developing cancer?
- • If so, should that be communicated back to the patient’s family? Can it?
It is certain that clinical annotation and risk assessment will be more costly and time-consuming than whole-genome sequencing.
Interpretation of Complex Genomes
Let’s face it, people, even with thousands of samples and accurate genotype information for millions of SNPs, we’re still struggling to suss out the genetic underpinnings of most common diseases. Just last week, I heard about the whole genome sequencing of a family quartet in which the two offspring, monozygotic twins, had a neurological phenotype of likely genetic origin. Yet even after numerous fancy variant-calling and filtering approaches were applied, the researchers were unable to pinpoint a cause. We’ll undoubtedly hear dozens of stories like these as large-scale efforts to determine the genetic basis of inherited diseases (e.g. Mendelian disorders) get under way this year. Yes, with sufficient samples, precise phenotyping, and comprehensive variant detection, we will have the statistical power to detect small-effect changes associated with a given phenotype. But that’s association, not causation. High-throughput functional assays may be required to determine if a certain variant is the actual cause.
When it comes to coding regions of the genome, we have a number of tools at our disposal to evaluate the consequences of an observed variant. RNA-seq can tell us if the gene is expressed, and if both alleles are represented. Computational algorithms can determine the likelihood that the change is damaging to the protein. High-throughput proteomics can even assess the level of protein in the cell. We can do a lot to investigate coding variants.
I wish I could say the same about noncoding variation. With the recent availability of exome sequencing, we’ve all had the luxury of cherry-picking variants in coding regions because these are less numerous and easier to interpret. But the simple reality is this: the vast majority of genetic variation in humans lies outside the exons of protein-coding genes. Anecdotal examples tell us that noncoding variation is quite capable of exerting influence on a phenotype, though the effect may be quite subtle. We have a lot more to learn about noncoding DNA, and we’ll need to study up in order to correctly annotate and interpret the vast catalogue of genetic variation in human genomes.

I think techniques like DNaseI-seq and FAIRE-seq, perhaps combined with transcription factor motif information, will be a promising first-pass approach to get at the non-coding variation.
Yes, I agree with the previous comment, and its already being done…!