Contents: dbSNP Growth • Build 135 Stats • Variant Composition • Function Classes • SNPs and Indels • Coding/Noncoding Tiers
Less than a decade ago, the leading experts estimated that there were approximately 10 million SNPs in the human genome. Those were the early days of post-genome research, when “The SNP Consortium” was formed and began BAC overlap comparisons to routinely identify and report SNPs. Believe it or not, in my old lab there were binders full of paper records documenting the evidence for each newly discovered SNP. These variants were submitted to a central repository of human sequence variation hosted at NCBI, appropriately named dbSNP.
Growth of dbSNP
The database has grown substantially, already exceeding the 10 million mark by 2006:
I highlighted some of the key driving forces of this growth that I happen to know about. These include the “BAC overlap” project of the SNP Consortium and similar SNP discovery efforts (2001-2003), The HapMap Project Phases I (2003-2005) and II (2005-2007), the advent of next-generation sequencing, of course, and most recently the 1,000 Genomes Project. You probably noticed a few trends in the figure above:
- Less-frequent dbSNP updates. In 2003-2004 when the HapMap consortium direly needed new loci, dbSNP was updating almost every month. New build releases have slowed down considerably, probably because (1) they’re less critical, and (2) it’s a much bigger job.
- Overall, and quite obviously, there’s been a rapid increase in submissions over time, with some phases of near exponential growth.
- The relationship between submissions (blue) and unique refSNP clusters (red). You’ll note that dbSNP gets more and more submissions, of which a shrinking fraction are truly novel loci.
Still, by 2009, there were about 18 million unique SNPs, nearly twice the predicted number. And large variant discovery projects fueled by next-generation sequencing, such as the 1,000 Genomes Project were just ramping up.
The Current State: dbSNP Build 135
Downloading the dbSNP database is not for the faint of heart. Even for bioinformaticians, the file formats offered (ASN1?) are somewhat intractable compared to BED files. I prefer instead to wait until the excellent team at the UCSC Genome Browser Database releases their annotation tracks for dbSNP builds, which contain the necessary information in far more accessible formats. They have just done so for build 135, and I did some quick-and-dirty parsing to come up with some statistics.
dbSNP Variant Composition
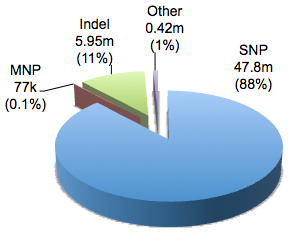
You might be surprised to learn that dbSNP contains not just SNPs, but several types of DNA sequence variation:
In the current build there are 54,212,076 unique variants with RS numbers, of which 47.8 million, or 88%, were single nucleotide polymorphisms. The remainder comprises insertion-deletion variants (indels, 11%), multiple nucleotide polymorphisms (MNPs, 0.1%), as well as ~420,000 other classes (named, mixed, and microsatellite). The named variants are old-school genetic markers (e.g. DS128384). Mixed polymorphisms are messy loci where multiple variant types (e.g. DNP and indel) are seen. Microsatellites, of course, are long stretches of repetitive sequences, such as di-nucleotide or tri-nucleotide repeats, whose length varies between individuals. Among these are the 15 short tandem repeats (STRs) utilized for forensic DNA profiling in CODIS, the FBI’s national DNA database.
dbSNP Function Classification
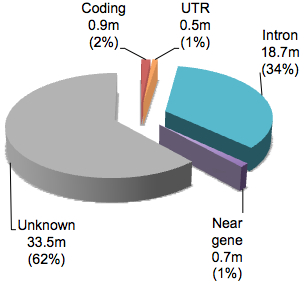
Variants in dbSNP are classified by their relationship to NCBI’s view of known protein-coding genes. There are about a dozen “function class” categories, but they can be grouped together into five types of sequences:
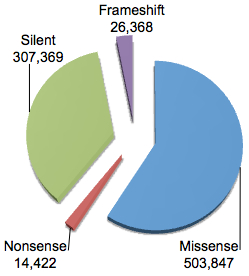
You will note that the vast majority have function classification of “Unknown” suggesting that these are non-coding variants not immediately adjacent from NCBI protein-coding genes. Even for variants in or around genes, 90% are classified as intronic. If we break down the variants that are in coding regions according to dbSNP:
You can see that the majority of coding variants (just over half a million) are classified as “missense”, meaning that they’re predicted to cause an amino acid substitution in the encoded protein. Most of the remainder are silent (synonymous), though there are also around 40,000 variants predicted to cause premature termination (nonsense) or a shift in translation frame (frameshift) in the encoded protein.
Honing in on SNPs and Small Indels
For next-generation sequencing analysis, I’m generally interested in two types of variation represented in dbSNP: SNPs and small (<50 bp) indels.
 The other types are either uncommon or too large to be readily detected with short reads, and further, there are curated, devoted databases that probably do a better job of representing them (e.g. Database of Genomic Variants for large indels and structural variants). Further, although the dbSNP functional classification is useful, we use an internal “tiering” system to represent variants according to their locations in the genome:
The other types are either uncommon or too large to be readily detected with short reads, and further, there are curated, devoted databases that probably do a better job of representing them (e.g. Database of Genomic Variants for large indels and structural variants). Further, although the dbSNP functional classification is useful, we use an internal “tiering” system to represent variants according to their locations in the genome:
- Tier 1 variants affect coding sequences, including exons, splice sites, and non-coding RNA genes
- Tier 2 variants occur in evolutionarily conserved or putative regulatory sequences
- Tier 3 variants are in non-coding, non-conserved, unique regions of the genome
- Tier 4 variants are in repetitive regions of the genome
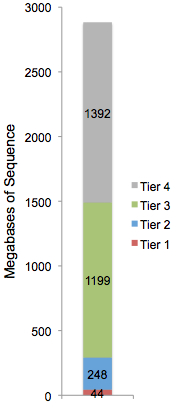
Every base in the reference sequence falls into one, and only one tier. Build 36 (hg18) of the human reference sequence is broken down to the right. There are 44 megabases of “tier 1” coding sequence in the human genome; that’s 1.53%, straight out of the textbooks. Tier 2 comprises 248 megabases, or 8.6%, which is slightly higher than the 5% expected rate of evolutionary conservation, probably because we’re fairly inclusive with what constitutes a putative regulatory element.
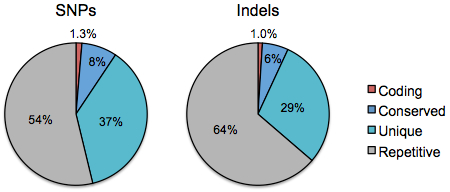
Distribution of SNPs and Indels by Tier
Next, we look at the distribution of dbSNP’s ~48 million SNPs and ~6 million small indels among the four tiers of genome space:
Strikingly, less than 10% of variants of both types fall into regions that are “interpretable” whereas the rest are in noncoding regions. The proportions of variants in tier 1 (1.3% of SNPs, 1% of indels) remains lower than the tier 1 fraction (above right), presumably due to purifying selection against changes to coding sequences. Many studies have shown this through far more careful analyses that account for ascertainment bias, population allele frequency, and other factors. It’s just fascinating to see the signature of natural selection in your basic pie chart.
I’m uncertain why the distributions in tiers 3 and 4 differ between variant types above, but there are likely a number of contributing factors. From a biological perspective, indels are both less frequent and subjected to greater natural selection than SNPs. From a technical perspective, SNP discovery algorithms are far more mature than indel discovery algorithms, owing in part to the difficulties of detecting the latter in relatively short sequence reads. We are currently, and have always been, better at finding SNPs than indels. With luck, the “accuracy gap” between SNPs and indels will diminish as sequencing technologies and detection algorithms continue to evolve.
References
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, & Sirotkin K (2001). dbSNP: the NCBI database of genetic variation. Nucleic acids research, 29 (1), 308-11 PMID: 11125122

Very nice. Quick question: your analysis of 135–is that just human?
Yes, that’s just human. Every organism tracked in dbSNP has its own build numbering; for example, mouse (Mus musculus) is currently on build 132 and soybean (Glycine max) is on 127.
Is the total chart also just human?
Yes, everything shown here is just for human.
Thanks. Quick question – what is a multinucleotide polymorphism (MNP)? I haven’t come across that term before.
How does it relate to a “haplotype”?
MNPs, according to dbSNP, seem to be substitutions of multiple adjacent base pairs. Dinucleotide polymorphisms, or DNPs, are a good example of these: you might see.
A haplotype is something else entirely – it’s essentially a set of variants that tend to be inherited together because they’re physically linked on the same chromosome. See the HapMap Project’s nice overview of the Origins of Haplotypes.
MNP, rs332 and rs333 are good examples.
http://snpedia.com/index.php/Rs332
http://snpedia.com/index.php/Rs333
Ack I take it back. Both are more correctly multi nucleotide indels, which are just indels under this scheme. please do not unscreen either of these comments.
The *shame* 🙂
However it might be worth mentioning the still nascent
http://www.ncbi.nlm.nih.gov/dbvar/
which is the NCBI equivalent of Database of Genomic Variants
I wonder however, how many of them are truly SNPs ? From my experience, and others, there are increasing numbers of somatic mutations (e.g. from cancer studies) and even RNA editing events in dbSNP, which shouldn’t be there. At the very least submitters should identify them as such.
Thank you for the nice analysis and graphics, and as a UCSC Genome Browser developer I’m very glad to hear that you’re finding our distillation of dbSNP useful!
I’d like to mention a common misconception about dbSNP. “Polymorphism” (the P in dbSNP) is a misnomer — a true polymorphism is a variant that occurs normally in a population, as opposed to a new mutation or rare disease variant, etc. dbSNP contains polymorphisms *and* any other short variants that are submitted to it, including known disease-causing mutations from locus-specific databases (i.e. databases devoted to particular well studied disease genes).
dbSNP recently changed their banner text to “Short Genetic Variants”, which is more accurate, but perhaps too subtle to be widely noticed. Unfortunately it’s too late for them to change their acronym! 🙂
Why does it matter that dbSNP contains disease mutations in addition to polymorphisms? — because many groups use dbSNP as a filter to remove “boring” variants from the large number of variants that they get from sequencing somebody’s genome. Not all dbSNP variants are boring! Using the entire dbSNP as a filter may result in throwing out some babies with the bathwater.
When it came to our attention that many groups (including a member of our scientific advisory board and some of our UCSC neighbors) were assuming that all variants in dbSNP are nonfunctional polymorphisms, we started thinking about extracting a subset of dbSNP — the truly boring variants — that would make a better filter. We came up with two subsets, which we call Common SNPs and Mult. SNPs (see tracks of those names in the Browser’s Variation track group: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&hgTracksConfigPage=configure+tracks+and+display#varRepGroup ). “Mult. SNPs” are variants whose flanking sequences map to multiple positions in the genome, which casts doubt on whether they are genetic variants — they might simply be slightly diverged duplications. By default, we now make those SNPs invisible in the All SNPs display. “Common SNPs” are uniquely mapped variants for which dbSNP has allele frequency data and whose minor allele frequency is at least 1%.
Out of 54,212,080 mappings in snp135, there are 3,538,479 multiple-mappers (snp135Mult) and 11,525,489 variants that match our criteria for “common” (snp135Common). That leaves a whopping 39 million uniquely mapping variants that might be rare or might simply be lacking allele frequency data. So filtering out our Mult. and Common subsets might be too weak of a filter, but in my personal opinion (not representing my employer here) that is still preferable to filtering out any variant that appears in dbSNP.
Last but not least, thanks to dbSNP for performing this gargantuan task and providing such a great resource!
Interesting post.. the plot charts are indeed helpful especially to look at the growth of validated SNPs vs the submitted SNPs. Considering the SNPs that are submitted and part of those that are validated how valid is it to say that a SNP that is in dbSNP is a SNP indeed and not a variant…
@Angie Hinrichs your insight on the two subsets is helpful.
How did you build the first chart? I mean, where did you get the data from?
Great question. I got the numbers from viewing the dbSNP summaries of each dbSNP release. I also noted the date of the release, and then I merged that into a new table.