New, smaller sequencing instruments are finding niches everywhere. Earlier this year in Less Is More: Sequencing on the Benchtop, I profiled three pint-sized sequencing instruments designed for next-gen sequencing on the benchtop: IonTorrent’s PGM, the Illumina MiSeq, and 454’s GS Junior. I must admit that I’m most intrigued by the PGM, probably because the idea of semiconductor sequencing is just so fascinating.
One concern that I’ve heard about the PGM, however, was sample preparation. The time requirement and difficulty of sample prep was an immediate question at AGBT when Jonathan Rothberg first presented the instrument. He dodged the question, and I don’t think there’s been a satisfactory answer since. Until now. This morning, IonTorrent announced a new companion instrument, the Ion OneTouch System, that streamlines sample preparation for the PGM sequencer. 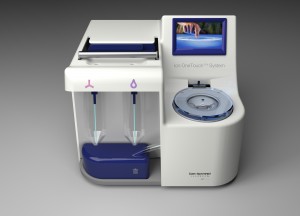
Priced at under $5,000, and measuring 1′ x 1′ square, the OneTouch requires “five minutes of hands-on time” and takes a few hours to run. It’s no longer the bottleneck, since the actual sequencing takes longer. The instrument is compatible with all three IonTorrent chips, which deliver throughputs ranging from 10 Mbp to 1 Gbp, with a single microgram of DNA.
PGM Applications
It’s a good thing to see the PGM constantly evolving – first the throughput was doubled, and now sample prep time cut substantially. At the price points we’re talking about, this might easily become standard equipment or small labs, academic departments, even single investigators. The current throughput of 1 Gbp isn’t enough for whole-genome or whole-exome sequencing, but it opens the door to a number of targeted applications. In Genome Technology’s Cancer Issue this month, for example, I read about a group that’s using the PGM for a clinical test comprising 100 common mutations in human cancers.
Essentially, the niche for PGM, MiSeq, and GS Junior is everything that’s not quite enough for a full-on sequencing run. A few examples come to mind:
- Microbial sequencing. For bite-sized genomes, 1 Gbp should be more than enough. Imagine walking into a clinic to have your strain of Streptococcus or some other infection sequenced the same day.
- Family linkage studies. With a few family members and a reasonably-sized linkage peak, you could sequence all gene-coding exons across a region of interest, either by PCR or custom capture.
- Orthogonal validation. Whole-genome and whole-exome studies might identify hundreds of putative mutations. Emphasis on putative. No matter how good your algorithms and filters are, there will be some false positives. Here’s an opportunity for a small, fast validation instrument. Preferably, you choose a different sequencing technology for validation (e.g. PGM for Illumina, MiSeq for SOLiD).
There are countless other potential applications; undoubtedly, investigators already have a few in mind. Access to high-throughput sequencing is getting less and less expensive. Now, you can have your own setup, for less than the price of a luxury sedan.
For more, see Keith Robison’s post at Omics! Omics! or Matthew Herber’s blog on Forbes.com.
I believe the price of the unit is under $50,000, not $5,000.
Your previous post stated the price would be under $50,000. Is $5,000 the correct amount?
Dan and Joseph,
Thanks for the question – the sequencer is $50K. The OneTouch, a separate instrument, is $5K.
I would like to have a comparison of the data from single molecule sequencing to those of current Illumina methods
My point is that the single molecule sequencing method theoretically provides a possibility that high throughput sequencing could be done because it solved, or at least relieved the chemical thermodynamics issues during the sequencing, which cause inaccuracy of sequencing data. But without seeing real data, I am not sure if there will be any other problems.
The accuracy of DNA sequencing data largely depends on the purity of template molecule. In the traditional sequencing, the template is cloned and amplified. Sequencing primer is designed by computational program under optimized condition. In NGS, the chemical thermodynamics optimization is mostly lost. Therefore both “under-represented sequences” and “generic sequences” become problem and cause difficulties in the data analysis and extraction of information. Single molecule sequencing method provides hope of solving these issues. That is why I am looking forward to seeing the real data
As in DNA microarray methods,
Basically, I believe that the accuracy for the NGS that is based on “PCR amplification followed by sequencing”, at base-to-base level is hard to improve due to the. chemical thermodynamics during the processes of PCR and sequencing. Single molecule sequencing provides a potential of overcoming these problems. The comparison between IonTorrent and other existed NGS systems obviously makes nonsense because none is surely right or accurate. The genome sequence data by IonTorrent data should be compared to some existing known genome database (sequencing was done by traditional sequencing method) tells how accurate it is.
Traditional sequencing is not 100% reliable, however, is far better than the NGS technologies. That is the only reliable standard.