The traditional cancer paradigm is one of progressive disease, in which cells gradually accumulate genomic rearrangements and point mutations over years (or decades), resulting in incremental progression through a series of increasingly malignant stages. New research has challenged that model. Using next-generation sequencing, Stephens et al have characterized a phenomenon in which tens to hundreds of rearrangements occur in a one-time cellular crisis.
The study comes from a group (led by Michael Stratton, Andrew Futreal, and Peter Cambpell) at the Sanger Institute that has pioneered the use of massively parallel paired-end sequencing to identify structural rearrangements in tumor genomes. During a rearrangement screen of ten patients with chronic lymphocytic leukemia (CLL), they identified one patient whose pattern of rearrangements was quite striking.
Massive but Localized Chromosome Remodeling
Over forty rearrangements affected a single chromosome (4q). The copy number across this segment oscillated between 2 (diploid segments with normal heterozygosity) and 1 (haploid segments with LOH). Interestingly, the latter regions did not result from simple deletions. There were breakpoints and copy number changes for deletions, duplications, and inversions. Although the breakpoints were markedly clustered, they joined segments that normally were not proximal to one another. Some were normally megabases apart. It was as if the chromosome were broken up to pieces and put back in random order.
That, as it turns out, is probably what happened.
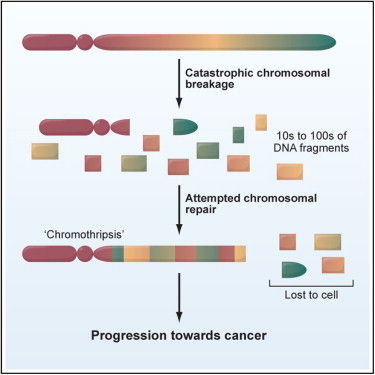
The patient was a treatment-naive, 62-year-old woman with CLL. She underwent chemo with alemtuzumab, but relapsed quickly. A sample of the relapse had the same rearrangements, but no new ones, suggesting that the process responsible for this phenomenon was not ongoing. Indeed, all evidence suggested that this was a one-time cellular crisis that the authors called “chromothripsis” – Greek for “chromosome shattering into pieces.”
The Chromothripsis Model, or Humpty-Dumpty
The proposed model suggests that a chromosome was literally broken into hundreds of pieces, likely while in its condensed state during mitosis. Humpty-Dumpty time. When this happened, the cellular DNA repair machinery (all the King’s men) responded, stitching the pieces back together as best it could. Some segments are rescued, assembled in random order (by non-homologous end-joining) into a derivative chromosome. Other segments are lost. This would explain the oscillating pattern of copy number and heterozygosity. Successfully “rescued” segments in the derivative chromosome would retain their heterozygosity and have a diploid copy number of 2. Lost segments would have LOH and a copy number of 1.
Recurrence in 2-3% of Human Cancers
To determine if this phenomenon was common in human cancers, the authors analyzed SNP array-based copy number profiles of 746 cancer cell lines. Of these, 18 (2.4%) exhibited the stamp of chromothripsis – frequent copy number changes in a localized region oscillating between one, two, and occasionally three states. Intriguingly, these cell lines represented numerous tumor types: melanoma, small-cell and NSC lung cancer, hematological malignancies, synovial sarcoma, and colorectal, esophageal, and thyroid cancer. Four cell lines were selected for paired-end sequencing and cytogenetics; only three are presented, though, because the fourth will be “described later” (apparently the authors plan to double-dip on publication).
A One-Off Cellular Crisis
Using the data from these three cell lines and some impressive Monte Carlo simulations, the authors demonstrate that the majority of rearrangements in each sample did not arise gradually over numerous cell divisions. Rather, they were the result of a massive but localized cellular crisis, one that likely occurred early in cancer development.
What could be the cause of such an event? The authors offer two interesting possibilities. A pulse of ionizing radiation, which is well known to cause double-stranded breaks, might have cut a swath through a chromosome condensed for mitosis. Another possibility is the breakage-fusion-bridge cycle that occurs with telomere attrition. Several of the observed chromothripses (?) extended into the telomere, which might support this explanation.
No matter how it happens, one expects that the vast majority of cells undergoing this crisis would die. A cell might survive, however, and the newly-rearranged chromosome could confer some advantages that promote malignancy – for example, loss of tumor suppressor genes, amplification of oncogenes, and dysregulation of gene expression. Chromothripsis, therefore, may provide a considerable leap towards cancer development.
Taken together, the results of this study suggest that 2-3% of all human cancers show evidence for tens to hundreds of structural rearrangements from a single catastrophic event. More studies are needed to understand the causes of this remarkable phenomenon, and the role it may play in cancer development and progression.
For more, have a look at Keith Robison’s very nice post on this study at his blog, Omics! Omics!
References
Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, & Campbell PJ (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell, 144 (1), 27-40 PMID: 21215367
This paper reports a phenomenon that has long been reported in array CGH data – called ‘firestorms’ – first appearing in Hicks et al. 2006 and many paper since.
Firestorms refer to complex rearrangements that are secluded to single chromosomes or chromosome arms. This paper is simply renaming ‘firestorms’ as chromothripsis and does not report anything new. Also the evidence for punctuated evolution is very weak, the data for temporal genomic kinetics is lacking.
This “chromothripsis” is actually not the same thing as “genomic firestorm” reported by Hicks et al. The many pieces of genomic fragments in chromothripsis osscillate between exactly (or mostly) two different copy number states, suggests the mechanism to be a one-off catastrophe, and then chromosome pieces glued together.
In the “firestorm” observed by Hicks et al, most of the cases involve high and complicated copy number variations, and Hicks et al suggest multiple, progressive mechanism.
Chromothripsis could also be categorized as a subset of “firestorm”, as long as detailed mechanisms are not considered. People just did not investigate deeply into them and did not realize that copy number pattern matters. However, in this chromothripsis paper, the authors definitely should mention and cite Hicks paper and the long known “genomic firestorm”. In the paper they just mentiond from 746 cancer cell lines they screened, there are 96 of them show clustered copy number changes, many of them caused by amplicons or “OTHER COMPLEX CLUSTERS OF REARRANGEMENTS”(without citation). Then, out of these 96, they found 18 lines to have what they dubbed CHROMOTHRIPSIS.