This week’s publication of the genome of a Korean individual in Genome Research marks the fifth individual whole genome sequenced with massively parallel sequencing platforms. The fact that this was not a Nature paper speaks as loudly as anything. The window of time when single whole genome sequences merit high-profile publications is slowly closing.
It is not terribly surprising, at least to me. At some point we are bound to reach a saturation, where the whole genome of a disease-free individual no longer yields novel information. The most tangible deliverable of such studies is a set of DNA sequence variants; without a phenotype there’s little relevance to human health, and with a sample size of 1 not much can be said about population genetics. Yes, there is some value in knowing the location and alleles of additional sequence variants, but look at dbSNP’s growth since 2003:
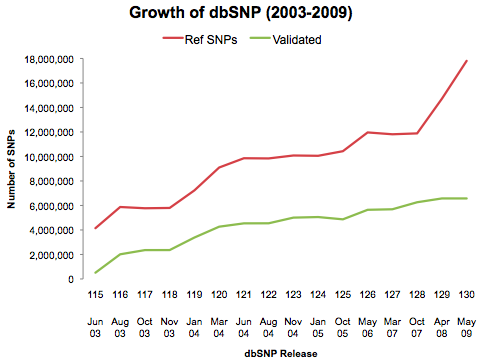
As of the last release there are over 17 million refSNPs (1 every 180 bp on average), where most experts agreed a few years back that there are probably ~10 million total in the human genome. The two rapid jumps in the plot above are the result of two massive NHGRI-driven efforts: The International HapMap Project (2003) and its sequel, the 1,000 Genomes Project (2008). Have we really not found all of the SNPs yet? Probably not. But as far as common SNPs – those prevalent enough to affect a sizeable fraction of the population – we must be getting close.
How Many SNPs Remain to Be Discovered?
The five genomes sequenced on massively parallel platforms to date, happily, have continental origins from Europe (2), Africa (1), and Asia (2). Yet despite this diversity of backgrounds, they all reported similar results for SNP discovery.
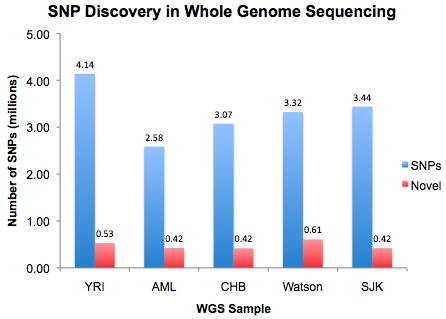 Note, for AML I’m only showing SNPs seen in both tumor *and* normal.
Note, for AML I’m only showing SNPs seen in both tumor *and* normal.On average, WGS studies identified 3.3 million SNPs per genome, of which ~479,000 (15%) are novel to dbSNP. That means about 1 out of every 1,000 bases yields a SNP, but only 1 out of every 6,700 bases yields a novel SNP.
The Future of Whole Genome Sequencing
As high-throughput genome selection technologies (e.g. capture) continue to develop, one begins to question whether or not WGS will retain its cache in the research community. My guess is that for “reference” samples (those without phenotypic information), it will be difficult to justify high-profile papers after the 1,000 Genomes Project. However, for disease and phenotype-related studies, especially cancer, I think that this will remain a hot topic. Even capture technologies rely on the basic assumption that the locations of functional variants are known (i.e. exons). That’s not always true, even for Mendelian disorders. Complex diseases are likely mediated by numerous loci (both coding and noncoding) interacting with many environmental factors. To puzzle these out, we probably will need to have the full genomes of affected individuals in hand.
References
[1] Wheeler, D., et al. (2008). The complete genome of an individual by massively parallel DNA sequencing. Nature, 452 (7189), 872-876 DOI: 10.1038/nature06884
[2] Ley, T., Mardis, E., Ding, L., et al. (2008). DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature, 456 (7218), 66-72 DOI: 10.1038/nature07485
[3] Bentley, D., et al. (2008). Accurate whole human genome sequencing using reversible terminator chemistry. Nature, 456 (7218), 53-59 DOI: 10.1038/nature07517
[4] Wang, J., Wang, W., et al. (2008). The diploid genome sequence of an Asian individual. Nature, 456 (7218), 60-65 DOI: 10.1038/nature07484
[5] Ahn, S., et al. (2009). The first Korean genome sequence and analysis: Full genome sequencing for a socio-ethnic group. Genome Research DOI: 10.1101/gr.092197.109
[…] a recent blog post at MassGenomics talking about the recently published sequence of a Korean individual, Dan Koboldt makes an […]