Working at the WashU Genome Center, I expect to encounter datasets that are large even by bioinformatician standards. But as we transition from traditional 3730-based sequencing to next-generation platforms, I’m beginning to appreciate just how much additional infrastructure we’ll need to handle the data flow. In the Medical Genomics group we’re constantly pushing up against capacity – servers, disk space, and man hours. None of these are in adequate supply for what’s ahead.
This is not to say that we’re without resources. In fact, the infrastructure already in place is considerable. We have about 500 computational servers (1600 cores) and nearly a petabyte (1,000 terabytes) of disk space. There’s an LSF system through which we submit and monitor jobs on The Blades.
You Didn’t Need That Done TODAY, Did you?
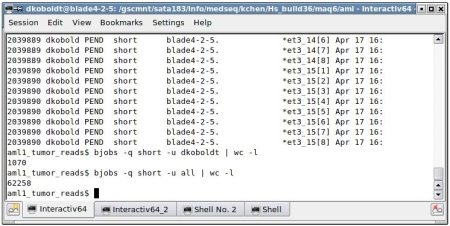
I submit about 1,000 small jobs and notice they’re all pending:
No doubt that’s because there are 61,000 jobs in front of me. We have a few different “queues” into which jobs can be submitted. The “short” queue is for jobs that execute in less than 15 minutes. At one job per core if every job finishes in 15 minutes, it looks like my jobs will start in about 9 hours. Oy.
The powers that be around here are rushing to build up our resources. As I’m not part of management, I can’t say for sure how long it will take to get the disk space and hardware we need. One thing I do know: we need a lot, and we need it soon.
This entry got picked up by Genome Technology By the way, they’re all through now.
$ bjobs -q short -u all
No unfinished job found in queue
Hi,
I would be interested to hear which algorithms are the bottlenecks in your NGS workflow.
Cheers
Thanks to the editors of Genome Technology’s Daily Scan, who picked up this post, saying, Somebody toss this guy a life preserver.