One significant challenges accompanied by advances in high-throughput genomic technologies (i.e. high-density arrays and next-gen sequencing) is that the speed at which we can detect candidate variants outstrips our ability to interpret their effect on phenotypes. In other words, genomic discovery is fast, but functional validation is slow. Also, the relationship between a sequence variant and an observable human trait tends to be complex, making it difficult to elucidate the mechanisms underlying an association.
Gene expression offers an appealing intermediate phenotype. It’s influenced by sequence variation, quantifiable, and can be assayed in high-throughput fashion. Early systematic efforts to identify expression quantitative trait loci (eQTLs) revealed some fascinating insights into the genetic regulation of gene expression, such as the fact that much of that genetic control occurs in noncoding regions. Also, most eQTLs were identified in proximity to the affected gene (cis-eQTLs), but some were millions of base pairs away or on other chromosomes entirely (trans-eQTLs).
Most eQTL surveys conducted thus far are limited to a certain cell type — most often, lymphoblastoid cell lines — and utilize genetic information from SNP arrays or low-pass whole genome sequencing (WGS). That’s why I’ve been excited to see the results of the Genotype-Tissue Expression (GTEx) project, which has applied RNA-seq to numerous different human tissues from donors that also underwent genome sequencing. In addition to a wide set of tissues profiled, their WGS data allows for a fairly comprehensive interrogation of genomic variation including large structural variants (SVs).
In the latest issue of Nature Genetics, members of the GTEx consortium led by several of my former WashU colleagues report the most extensive study to date of the impact of structural variation on gene expression. Their dataset comprises RNA-seq from 13 different tissues along with deep WGS (blood) from a total of 147 individuals.
Genome-wide Structural Variation
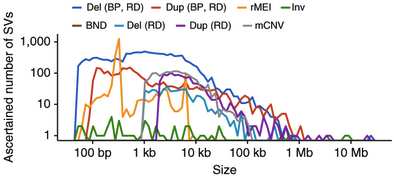
A strength of this study is that the authors have considerable expertise in identifying SVs from genome sequencing data. Across the 147 individuals, they identified 23,602 high-confidence SVs, including deletions, duplications, inversions, and other SVs.
Common SVs Affect Gene Expression
Of the 9,000 common SVs (MAF>0.05) in their dataset, 1,634 were associated with gene expression variation in at least one tissue. Because an SV can influence gene expression in multiple tissues and vice-versa, their complete analysis uncovered 5,128 individual eQTLs affecting 2,064 different genes.
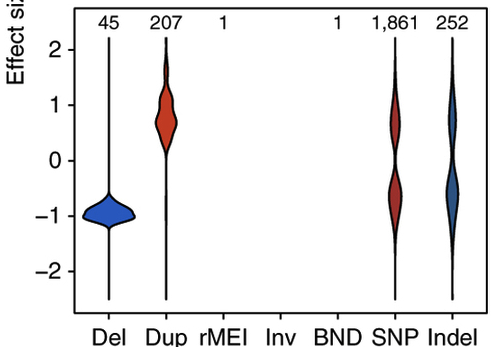
Some 11% of the eQTLs involved an SV that altered one or more exons of the affected gene. More than 90% of the time, the effect on gene expression is consistent with the type of SV involved, i.e. deletions tend to decrease expression and duplications tend to increase it:
That’s a nice result if not an altogether surprising one, since you’d hope that changing the genomic copy number of a gene would alter its expression in somewhat predictable ways.
Contribution of SVs to eQTLs
The authors also identified eQTLs involving SNVs and small indels, which are more numerous and generally easier to detect than SVs with current next-gen sequencing technologies. This offered a more complete picture of genomic variants affecting gene expression, and allowed them to characterize the relative contribution of each type of variant to eQTLs. Using fine-mapping approaches, Chiang et al found that for 3.5 to 6.8% of cases, a structural variant was the probable causal variant.

Heritability of each eQTL apportioned to the most significant SV in the cis window (X-axis) and the additive effect from the top 1,000 most significant SNVs and indels in the cis window (Y-axis) in joint eQTL (left) or mapping (right) analyses (Chiang et al, Nat. Genet. 2017)
This might seem like a trivial contribution, but remember that the typical human genome harbors ~4 million SNVs and indels, but just 5-10,000 SVs. On a per-variant basis, an SV was 28 to 54 times more likely to modulate gene expression than an SNV or indel. Moreover, this is probably an under-estimate of the contribution of SVs, since our ability to detect/genotype them with current WGS technologies is considerably less than our ability to detect smaller variants.
Noncoding SVs Affect Gene Expression
Interestingly, 88.3% of SVs predicted to cause an eQTL did not affect gene dosage or structure. Instead, they likely exert their effects via noncoding regulatory mechanisms. Cross-referencing them with several classes of predicted regulatory elements revealed that SV-eQTLs were significantly enriched in regions that were:
- 10kbp upstream or downstream of the gene transcript
- Within 1 kbp of an enhancer
- Predicted to bind transcription factors (by FunSeq)
Overall, this study beautifully illustrates the important role that structural variants have in modulating gene expression. It also lends additional evidence to what I’ve been saying for a long time: that noncoding regulatory variants probably underlie the majority of inter-individual differences in humans.