The real cost of sequencing is as hard to pin down as a sumo wrestler. Working in a large-scale sequencing laboratory offers an interesting perspective on the duality of the so-called “cost per genome.” On one hand, we see certain equipment manufacturers and many people in the media tossing around claims that sequencing a genome now costs under $1,000. On the other, we write grant budgets and estimates based on actual costs, which include things like sample assessment, variant calling, and data storage. With these incorporated, the cost per genome is not that low, even for large projects.
I came across a wonderful opinion piece at Genome Biology, in which the authors discuss the evolution of sequencing and computing technologies over the past 60 years. Admittedly, I found it a bit daunting at first, because theories of computation and “conceptual frameworks” don’t excite me. Once I pushed past the organizing principle stuff, however, I found it contained some shrewd perspectives on the current state and near future of genomics.
Big Data: Large Scale Sequencing
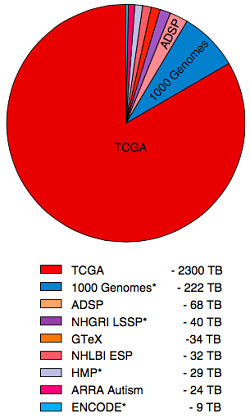
The rise of next-gen sequencing factors significantly in the big data paradigm for genomics. Rather than trot out the sequencing cost versus Moore’s law figure, the authors provided some compelling illustrations of the dramatic increase in the pace and quantity of sequencing. The most striking of these was a pie chart of the sequence data contributed by large-scale projects.
The Cancer Genome Atlas (TCGA) dwarfs everyone else, with 2300 Terabases of sequencing data. This is ten times the amount generated by the 1,000 Genomes Project, and 30 times the amount in the Alzheimer’s Disease Sequencing Project (ADSP).
Costs and Economies of Scale
A key concept highlighted by the authors is the interplay between fixed and variable costs. The sequencing technologies utilized for the Human Genome Project had considerable up-front costs (i.e. instrument purchase) and somewhat fixed per-sample costs. In contrast, next-generation sequencing has a high up-front cost, but a reduced per-sample cost as volume increases. In other words, the more genomes we produce, the less they cost. True, this economy of scale has an upper limit, but the current throughput of an Illumina X Ten system — 18,000 human whole genomes per year — provides enormous capacity.
Interestingly, the opposite paradigm-shift is taking place in the computing industry. Until recently, the model for computing mirrored NGS: large up-front cost of buying the servers, but lower variable costs for running them. In some ways, this erected a barrier for smaller labs hoping to tackle complex problems, because they might not be able to afford enough computing equipment to handle the workload. Yet cloud computing and computing-as-a-service platforms have largely removed the need for that up-front investment. Anyone can buy as much computing power as they need on the Amazon or Google clouds. Although the variable cost (per CPU hour) is higher than that of a large data center, there’s no large fixed cost at the front end. As the authors put it:
This new regime, in which costs scale with the amount of computational processing time, places a premium on driving down the average cost by developing efficient algorithms for data processing.
As a bioinformatician, I think this is a good thing, because it forces us to improve our software tools and pipelines to become as efficient as possible.
Although cloud computing offers tremendous appeal, it faces some challenges for widespread adoption in our field. Most sequencing take place in academic settings, where equipment purchases are often exempt from indirect fees (because the university can write off depreciation). Also, many investigators don’t have to pay for the basic utilities required to run computing equipment (e.g. electricity and cooling). These factors encourage us to stick with the traditional computing model, rather than shifting to cloud computing which will be subject to indirect costs.
Breaking Down the Cost of Sequencing
We tend to measure the cost of sequencing as bases per dollar, or more recently, X dollars per genome. Both funding agencies and sequencing customers like to ask how much an exome or a genome costs. This single-price figure has some disadvantages:
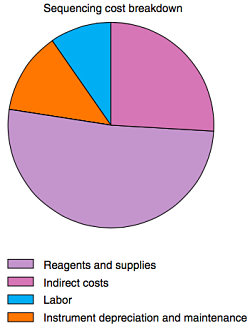
- It’s not always clear what that dollar figure includes. Is it purely the sequencing run cost, or does it account for non-free things like sample assessment, handling, and bioinformatics analysis? Notice how they’re not included in the figure at right.
- It obscures the true cost breakdown of a sequencing project into its constituent parts, which complicates cost estimates and makes it harder to adapt to changes like the shift to cloud computing.
- It can lead to unrealistic expectations. People hear about this $1,000 genome, so they come to us for a whole-genome sequencing quote, and get upset when (1) it’s not that low, and (2) we have to add other costs, like sample handling, to the estimate.
Unrealistic expectations are a source of constant frustration for us. When we provide estimates for a sequencing project, we include analysis time as a recommended (but often not required) line item. Of course, no one wants to pay for analysis — they just want the sequencing. Sometimes this is just fine — we provide sequencing for a number of collaborators who are capable at NGS analysis. Other times, the customer later asks “How do I open this BAM file to see my variants?”
Sorry, but high-quality variant calls require analysis, and as I’ve written before, bioinformatics analysis is not free.
One thing that concerns me about the current state of federal funding (for sequencing) in the United States is that large-scale projects emphasize data production, not data analysis. The RFA for NHGRI’s large-scale sequencing program (CCDG) mandated that 80% of the budget go to data production. Yet as the authors of this opinion piece correctly point out:
As bioinformatics becomes increasingly important in the generation of biological insight from sequencing data, the long-term storage and analysis of sequencing data will represent a larger fraction of project cost.
I couldn’t agree more.
References
Muir P, Li S, Lou S, Wang D, Spakowicz DJ, Salichos L, Zhang J, Weinstock GM, Isaacs F, Rozowsky J, & Gerstein M (2016). The real cost of sequencing: scaling computation to keep pace with data generation. Genome biology, 17 (1) PMID: 27009100