Three letters to Nature Genetics published (online) this week report the association of a rare nonsynonymous variant in the C3 gene with risk for age-related macular degeneration (AMD). Full disclosure: I’m an author on one of them. These studies all came from different groups, working primarily with different case-control cohorts. What’s fascinating is how they all arrived at the same answer, at around the same time, but in very different ways. They also shared a starting point, of sorts: the relatively recent association of variants in complement pathway genes with risk for AMD.
The Complement System
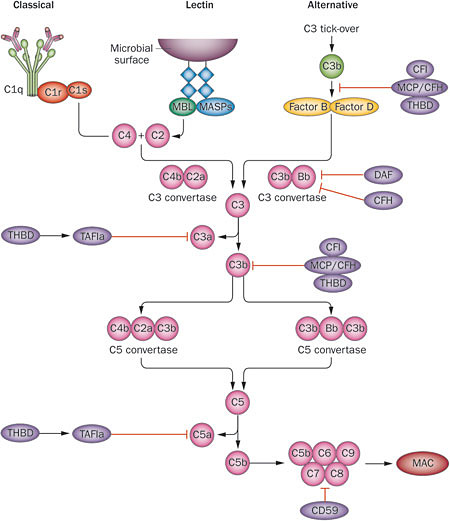
The complement system, sometimes called the “complement cascade”, is a complex network of plasma (blood) proteins that cooperate to provide microbial defense and maintain healthy tissue. It’s part of the immune system, and complements the antibody- and cell-driven responses to infection by pathogens. Thus the name. There are three ways that the complement system becomes activated:
- The classic complement pathway is triggered by antigen-antibody complexes, which bind a protein complex named C1. This induces a conformational change, causing C1 to activate two other complexes, C2 and C4. These are cleaved and form C3 convertase, which promotes cleavage of C3 into C3a and C3b. The latter becomes part of C5 convertase, and it goes on from there (see below).
- The lectin pathway is triggered by mannose residues found on bacterial membranes, which are bound by mannose-binding lectin (MBL), but otherwise is quite similar to the classic complement pathway.
- The alternative complement pathway is unique because it’s always activated at a low level, and doesn’t require a microbial surface to trigger it. Instead, it’s triggered by endogenous C3 proteins, which sort of float around until they stick to something. Regulatory molecules keep C3 off of healthy host cells. However, it gradually accumulates on pathogen surfaces and “altered self” cells, eventually activating the alternative pathway.
Activation of the complement system eventually culminates in the formation of the membrane attack complex (MAC). This is the tip of the spear of the complement system; it literally punches holes in target cells, causing them to swell and die. Such a potent response obviously might pose a hazard to the host as well, so there are numerous regulatory molecules that act as negative regulators of complement activation. Of particular interest are complement factor H (CFH) and factor I (CFI), which promote the cleavage of C3b to its inactive form.
Importance of the Complement System
The intricacy and importance of the complement system is evidenced by inherited complement deficiencies. Mutations in the terminal (cell-destructive) portion of the complement pathway knock out its effector function, and causes susceptibility to infections by gram-negative bacteria. In contrast, mutations affecting complement regulatory proteins have been linked to auto-immune conditions like systemic lupus erythematosus. More recently, genetic studies of AMD have suggested that variants in C3, CFH and CFI might influence susceptibility to this disease. It’s an intriguing proposition, in particular because one could imagine a late-onset phenotype resulting from dysregulation of the alternative complement pathway. That’s where the three studies published this week got started.
Three AMD Studies
The deCODE Study
Helgason et al have performed low-pass whole-genome sequencing (10x) on 2,230 Icelanders, which enabled them to impute genotypes for 34.2 million variants in 1,143 cases and 51,435 controls. Of the 12 established AMD loci, the authors found genome-wide significance for six (APOE, ARMS2-HTRA1, C2-CFB, CFH, C3, and TIMP3). Using regression analysis, the authors identified the best association signal at each locus, finding that for 5/6 loci, it matched the most significant finding from a recent meta-analysis. The sixth locus, C3, had a common variant hit corresponding to known associated variants p.Arg102Gly and p.Pro314Leu. But there was also a rare missense variant (p.Lys155Glyn, rs14785925) that was significantly enriched in cases. It was independent of the known risk variant in CFH (p.Arg1210Cys). Follow-up genotyping confirmed Lys155Gln in 1,107 cases and 2,869 controls from the Icelandic cohort. Replication in two independent cohorts (1,525 cases, 1288 controls from the U.S. and 1,329 cases, 1,689 controls from Europe) upheld the hit; the combined dataset yielded an odds ratio of 3.65 with a p-value of 8.8e-16. Given the role of C3 in complement activation, and the observation that all three associated variants (the two common hits and this one) are in the C3b portion, the authors speculated that reduced binding between C3b and CFH might lead to increased C3 convertase activity.
The AMD Targeted Sequencing Study
Our group undertook an effort to systematically identify rare, large-effect variants by targeted sequencing of eight AMD risk loci identified by GWAS. We performed custom capture and Illumina sequencing of 57 genes in those loci in 3,124 individuals (2,335 cases, 789 controls) recruited at the University of Michigan and the University of Pennsylvania as part of the Age-Related Eye Disease Study (AREDS) sponsored by the National Eye Institute. To increase power, we selected ancestry-matched controls from the NHLBI Exome Sequencing Project cohort, yielding a final discovery set of 2,268 cases and 2,268 controls. This analysis revealed two large-effect rare variants: the previously reported variant (p.Arg1210Cys) in CFH and the newly identified p.Lys155Gln variant in C3. Follow-up genotyping in 4,526 cases and 3,787 controls from the U.S. and Europe replicated this finding. Further, by genotyping 471 families with multiple AMD cases, we estimated that 75% of first-degree relatives of a p.Lys155Gln carriers who also had AMD would carry the variant, consistent with an odds ratio of ~3.
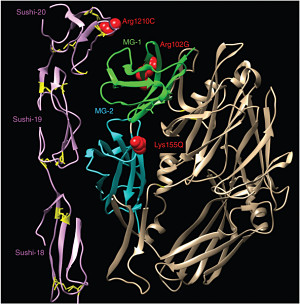
We investigated the possible functional consequences of p.Lys155Gln through in silico modeling of the protein crystallography data, which revealed that CFH variant p.Arg1210Cys, C3 variant p.Lys155Gln, and C3 variant p.Arg102Gly all map near the surface where CFH and C3b interact, suggesting that they might affect binding.
The Seddon/Raychaudhuri Sequencing Study
The authors of the third study sequenced the exons of 681 genes within all reported AMD loci and related pathways in 1,676 cases, 745 controls, and 36 siblings with discordant disease status. A burden test of rare (f < 0.01) protein-altering variants revealed that 7.8% of AMD cases are carriers of rare missense variants in CFI, compared to 2.3% of controls. Single-variant analysis uncovered 5 risk and 15 protective variants with evidence of association, of which four were within or near CFH (including p.Arg1210Cys). The 11 variants outside of CFH were genotyped in 2,227 cases and 2,888 controls from Boston, Baltimore and France. Two rare, replicated variants emerged: p.Pro167Ser in C9 and p.Lys155Gln in C3. Seddon and colleagues noted the location of the C3 variant close to the binding site for CFH, reasoned that the p.Lys155Gln substitution in C3 might confer resistance inactivation by CFI (which is aided by CFH). Indeed, fluid-phase cofactor assays demonstrated a significantly reduced cleavage of C3 with Gln compared to the wild-type (Lys) at residue 155.
Complement Mutations and aHUS
Interestingly, uncontrolled complement activation has long been implicated in hemolytic uremic syndrome (HUS), a severe blood disorder that primarily affects children. Usually, HUS is caused by an infection from shiga-like toxin-producing E. coli HUS (STEC-HUS) or streptococcus pneumoniae (pneumococcal HUS). A rarer and more severe form called atypical HUS is caused by mutations in complement genes (C3, CFH, CFI, and others) that result in chronic complement activation. I took a moment to look in OMIM, and found that some of the key variants we’re discussing (p.Arg1210Cys in CFH, p.Arg102Gly in C3) were reported in aHUS patients. There’s a lot of overlap. Yet HUS as a whole is relatively rare (21 cases per million persons per year) and affects mostly children, whereas AMD is relatively common (212 cases per million persons per year) and affects older adults. Clearly there are other factors (genes, pathogens, environment) involved in these diseases. So there’s still work to do. But taken together, these three studies demonstrate the power of high-throughput genotyping and sequencing technologies to uncover rare variants contributing to disease risk, and have shed more light in the role of alternative complement activation in AMD risk. References Helgason H, Sulem P, Duvvari MR, Luo H, Thorleifsson G, Stefansson H, Jonsdottir I, Masson G, Gudbjartsson DF, Walters GB, Magnusson OT, Kong A, Rafnar T, Kiemeney LA, Schoenmaker-Koller FE, Zhao L, Boon CJ, Song Y, Fauser S, Pei M, Ristau T, Patel S, Liakopoulos S, van de Ven JP, Hoyng CB, Ferreyra H, Duan Y, Bernstein PS, Geirsdottir A, Helgadottir G, Stefansson E, den Hollander AI, Zhang K, Jonasson F, Sigurdsson H, Thorsteinsdottir U, & Stefansson K (2013). A rare nonsynonymous sequence variant in C3 is associated with high risk of age-related macular degeneration. Nature genetics PMID: 24036950 Zhan X, Larson DE, Wang C, Koboldt DC, Sergeev YV, Fulton RS, Fulton LL, Fronick CC, Branham KE, Bragg-Gresham J, Jun G, Hu Y, Kang HM, Liu D, Othman M, Brooks M, Ratnapriya R, Boleda A, Grassmann F, von Strachwitz C, Olson LM, Buitendijk GH, Hofman A, van Duijn CM, Cipriani V, Moore AT, Shahid H, Jiang Y, Conley YP, Morgan DJ, Kim IK, Johnson MP, Cantsilieris S, Richardson AJ, Guymer RH, Luo H, Ouyang H, Licht C, Pluthero FG, Zhang MM, Zhang K, Baird PN, Blangero J, Klein ML, Farrer LA, Deangelis MM, Weeks DE, Gorin MB, Yates JR, Klaver CC, Pericak-Vance MA, Haines JL, Weber BH, Wilson RK, Heckenlively JR, Chew EY, Stambolian D, Mardis ER, Swaroop A, & Abecasis GR (2013). Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nature genetics PMID: 24036949 Seddon JM, Yu Y, Miller EC, Reynolds R, Tan PL, Gowrisankar S, Goldstein JI, Triebwasser M, Anderson HE, Zerbib J, Kavanagh D, Souied E, Katsanis N, Daly MJ, Atkinson JP, & Raychaudhuri S (2013). Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nature genetics PMID: 24036952