It might surprise you to learn that the majority of genes found in human beings are not our own — they belong to the hundreds of species of bacteria that make up the gut microbiome. Just as the number of human genomes sequenced on next-gen instruments has grown exponentially in recent years, so too have the number of these “metagenomes”. And that’s a good thing, because the gut microbiome plays an important role in human health and has been linked to a growing number of diseases.
Framework for Metagenomic Variation Analysis
A new study this week in Nature describes the genomic variation landscape of the human gut microbiome, derived from sequencing and analysis of 252 metagenomes from 207 individuals. This is a far more complex challenge than analyzing a few hundred human genomes; it’s what many refer to as “metagenomics” – sampling and sequencing the genomes of a number of species. The gut microbiome comprises hundreds of different species, many of which are related, but each with its own reference genome. One of the major achievements of this study was a comprehensive framework for the analysis of metagenomic data:
- The authors generated a set of references from 1,497 prokaryotic genomes, which clustered (on the basis of 40 species-identifying genes) into reference genomes for 929 species.
- They aligned metagenomic data data from 207 individuals — representing U.S. (NIH Metagenome and WashU) and European (MetaHIT) cohorts — for a total of 7.4 billion mapped metagenomic sequence reads (42% of the total) averaging 80 bp in length.
- Finallly, they isolated 101 prevalent microbe species (>40% of reference genome covered) and took these forward for genomic variation analysis.
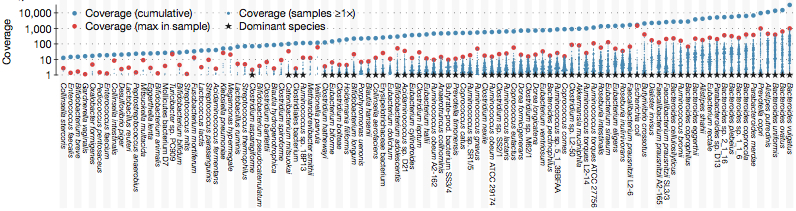
Composition of the Gut Microbiome
The minimum coverage requirement was 10x (cumulative) for the prevalent species; actual base pair coverage among prevalent species ranged from 12x to 32,400x:
To enable more accurate comparative analyses, the authors employed multi-sample calling of SNPs, small indels, and SVs across metagenomic samples.
Genomic Variation in the Gut Microbiome
To enable more accurate comparative analyses, the authors employed multi-sample calling of SNPs, small indels, and SVs across metagenomic samples among the 101 prevalent microbe species. They identified 10.3 million SNPs, an impressive number given that the entire reference comprised 329 Mbp. It’s nearly as many, in fact, as the ~14 million SNPs reported for 179 human genomes. And our reference is about ten times larger, so clearly the nucleotide diversity in our gut microbes far outweighs that in our constitutional genome.
There were other forms of genetic variation as well: 107,991 small insertion/deletions (indels) and 1,051 structural variants (SVs) detected using Pindel. As we’ve observed in other genomes, indels occur far less frequently than SNPs. Empirically, in human genomics we expect about a 1:10 ratio of indels to SNPs, and that holds true here (108K indels, 10.3m SNPs).
Evolution and Selection
The rigorous analysis framework and large number of SNPs enabled the authors to compare, for the first time (at this scale), the evolution of different coexisting species across a large cohort of individuals. To do so, they calculated the the ratio of nonsynonymous to synonymous polymorphisms (pN/pS) within each species in every sample. This ratio was 0.11 on average, and remained stable at coverages higher than 10x (a good indication of accurate SNP calling), but varied considerably among species.
Focusing on the 66 most dominant species, the authors found that relatively low pN/pS ratios were constant across different hosts, which may indicate similar selective constraint across individuals. This implies that the evolution of gut microbiota is likely dominated by long-term purifying selection and genetic drift, rather than rapid adaptation to new host environments. Further, the low pN/pS ratio observed for genes related to type IV secretion systems suggests that interaction with the host’s immune system is under purifying selection, and maintaining genome plasticity (as well as antibiotic resistance) is essential for gut species.
Individuality and Temporal Stability
There were 88 individuals from the US cohort that were sampled at different time periods (~1 year) with no antibiotics in between. Looking at the composition and genomic variation of these gut microbiomes, the authors found that while species abundance changed over time within an individual, the variation patterns were remarkably stable. In other words, healthy individuals retained (for a long term) specific strains of microbiota. Also, there were no clear geographical differences between European and U.S. cohorts. All of this suggests that every human has a unique metagenomic profile that’s stable over time. Almost like a fingerprint.
It won’t be long before CSI shows and IRBs jump on this source of potentially identifiable information. So be careful where you, you know… go.
There’s much more to this work than I can cover here, including studies of variation and selection in key bacterial genes. Moving forward, the authors have provided a framework that should enable even larger-scale surveys of the human gut microbiome.
References
Schloissnig, S., Arumugam, M., Sunagawa, S., Mitreva, M., Tap, J., Zhu, A., Waller, A., Mende, D., Kultima, J., Martin, J., Kota, K., Sunyaev, S., Weinstock, G., & Bork, P. (2012). Genomic variation landscape of the human gut microbiome Nature DOI: 10.1038/nature11711