Next-generation sequencing is a disruptive technology. It’s changed the way we conduct genetic and genomic research, and perhaps more importantly, where that research takes place. Because high-throughput sequencing is accessible not just to large genome centers but to the wider research community. With this “democratization” of sequencing, many things are changing. Virtually any investigator can conduct exome or whole-genome sequencing, if not in their own lab, then with assistance from a service provider.
As I’ve written before, samples are the new commodity. And as we enter an era of pervasive sequencing, there are significant challenges ahead.
Human Genetics Sample Challenges
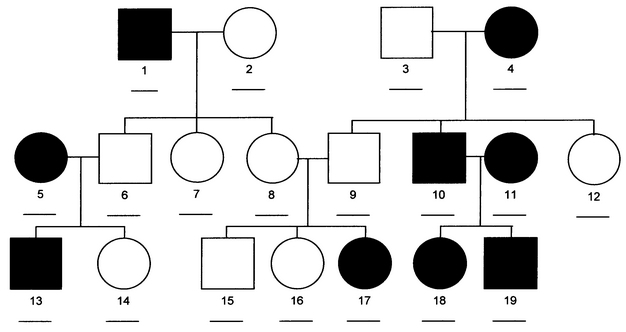
Exome sequencing promises to help elucidate the genetic basis of many rare inherited disorders. Indeed, NHGRI recently funded several groups to tackle Mendelian diseases with exome sequencing as one of the primary tools. Dozens of disease-causing mutations identified by exome sequencing in the past few years have demonstrated that this approach is fruitful for many such disorders.
One problem, however, is that Mendelian diseases, particularly those in which the causal mutation remains unknown, tend to be exceptionally rare – often less than 1 case in 10,000 or even 100,000 births. Simply collecting enough samples to even undertake an exome sequencing study is difficult. Given the low barrier to entry for exome sequencing these days, there are undoubtedly many research groups competing for the same samples.
The most powerful Mendelian disease studies are facilitated by large family pedigrees with multiple affected individuals (and unaffected but related controls), all of whom:
- Have been diagnosed (or cleared) by a specialist using standardized criteria
- Are alive and capable of providing a sample
- Are willing to provide a sample
- Agree to sign ever-more-frightening consent forms
Unfortunately, a substantial fraction of family cohorts won’t meet all of these criteria. Crucial family members may have passed away, or moved elsewhere, or simply have no interest in providing a sample. Growing concerns about individual privacy and sample misuse undoubtedly reduce volunteerism. Heck, I’m in the field myself and I’d be nervous about granting someone full access to my genetic information.
Even if you can get the samples to study a disease of interest, you have to get the right samples. Recent reports of somatic mosaicism — in which the causal mutation in a patient is only present in certain tissues — have highlighted that a blood or saliva sample might not be enough.
Cancer Genomics Sample Challenges
Few fields of research have benefited more from next-gen sequencing than that of cancer genomics.
The genomes and exomes of thousands of human cancers have been characterized already, implicating new genes and pathways, informing diagnosis and prognosis, and highlighting new therapeutic targets. The more we’ve sequenced cancer genomes, the more we’ve come to realize how complex they are:
- Many tumors comprise multiple, genetically distinct sub-populations of cancer cells (subclones), each with a different complement of somatic alterations.
- Tumor cells constantly undergo mutation and evolution to compete for growth advantages, to metastasize, to avoid host defenses, and to survive therapy.
- The vast majority of somatic mutations in a tumor genome are unique to that genome. Few are shared (or “recurrent” as we like to say) between tumors from different patients, even when hundreds of tumors of the same type are screened.
One of the collaborating oncologists I work with pointed me to a recent protocol review on measurement of prognostic factors in breast cancer. In it, the authors acknowledged the growing array of tools and technologies for studying human breast tumors, but noted that “Routine pathologic evaluation of a breast cancer must never be compromised by the demand to submit portions of the tumor for special studies.”
There are many, many studies that could be performed on a tumor specimen, but there’s only so much tumor to go around. Further, when patients undergo treatment prior to the surgery that removes the tumor (this is called neoadjuvant therapy), the goal is to shrink the tumor. That makes for even less material to study, and now you’re working with a tumor that has been influenced by chemotherapy.
Fewer Samples and Less DNA
One interest of many technology development groups is reducing the amount of input DNA (or RNA) required for capture and/or sequencing. My interpretation of their results is simple: the more DNA, the better. And yet for the reasons I’ve outlined above, we’re likely to see less and less available DNA for sequencing experiments.
Whole-genome amplification (WGA) seems like a reasonable strategy to obtain enough DNA for sequencing. The problem is that WGA tends to produce artifacts that look like low-frequency variants. One can eliminate many of these, but they’re virtually indistinguishable from, say, somatic mutations unique to a minor clone in a tumor sample.
Immortalized cell lines, such as those created for the HapMap and 1,000 Genomes Projects, provide plenty of high-quality DNA. As much as you want. Unfortunately, cell lines acquire some random mutations during the immortalization process. Not many, but enough that those well-characterized trios are virtually useless for discovering true de novo mutations.
Formalin fixation and paraffin embedding (FFPE) has been the standard of sample archiving among pathologists for decades. There exist, right now, large collections comprising thousands of well-characterized tumors dating back 20 years or more. There’s treatment response and relapse and other outcome data. But there are a couple of drawbacks to these incredible resources. First, the process of FFPE generates artifactual random mutations due to formalin cross-linking of cytosines on either strand, leading to misincorporation of adenosines during PCR. Also, the matched normal tissue for these samples may not be available, making it impossible to determine if a mutation is truly somatic.
Sample Ethics and Propriety
A growing number of ethical and legal issues surrounding human samples and DNA sequencing have become more prominent in recent years. The recognition of how valuable certain samples are becoming has already caused legal spats between investigators and their institutions. Who owns the sample? The clinician or his employer? In the future, access (and rights) to a relevant cohort will be an even more important part of grant applications.
There are many ethical angles to consider, enough that I hesitate to scratch the surface. I’ll mention two concerns: first, one of privacy. As we learn more about genetic variation and its role in disease, the predictive power (and perceived predictive power) of genetic information grows. What if that gets into the wrong hands?
I know, we have GINA legislation. Still, one could easily picture a nefarious corporation that, when interviewing potential employees, collects the candidate’s empty soda can and puts it into a clear plastic bag for quiet testing with a loyalty-productivity-intelligence-longevity SNP chip.
For any patient considering an informed consent document, there are uncountable considerations. Because your genetic information does not just belong to you. Thanks to bioinformaticians like us, when you grant access to your genome or exome data, you’re not just revealing things about you. We’ll learn about your parents and grandparents. We’ll know something about your children or future children.
A sample’s not just a sample any longer. It’s a family legacy.
References
Yost SE, Smith EN, Schwab RB, Bao L, Jung H, Wang X, Voest E, Pierce JP, Messer K, Parker BA, Harismendy O, & Frazer KA (2012). Identification of high-confidence somatic mutations in whole genome sequence of formalin-fixed breast cancer specimens. Nucleic acids research, 40 (14) PMID: 22492626