Human cancers are driven by genetic and epigenetic changes to the genome of healthy cells. We often think about acquired mutations as the key drivers of tumor development and growth. Most studies empowered by next-gen sequencing have focused on identifying these changes. Yet aberrant DNA methylation – hypermethylation of tumor suppressor genes and hypomethylation of oncogenes – is now recognized as a hallmark of cancer cells. A new study in Genome Research describes one of the first efforts to survey DNAm genome-wide in a human cancer type.
Neurofibroma and Peripheral Nerve Sheath Tumors
Feber et al employed a relatively new application of NGS, called methylated DNA immunoprecipitation sequencing (MeDIP-Seq), to identify differentially methylated regions in malignant peripheral nerve sheath tumors (MPNSTs). The precursor to MNST is neurofibroma, a disease of largely benign tumors that arise from normal Schwann cells.
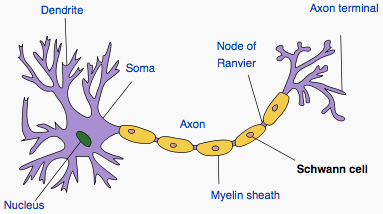
Schwann cells wrapped around an axon (Wikipedia)
Most neurofibromas arise in patients with neurofibromatosis type 1, an autosomal dominant disorder caused by germline mutations in the NF1 gene. There are also sporadic cases of NF, presumably due to acquired mutations in NF1. Neurofibroma affects 1 in 3,000 individuals worldwide; 5-10% of these patients go on to develop MPNST, which has a 5-year survival rate of <50%. Why do some patients progress when others do not? Aberrant DNA methylation offers an intriguing possibility.
Pooled Samples and MeDIP-Seq
The authors thus performed comparative DNAm analysis of three types of samples:
- DNA from 10 malignant peripheral nerve sheath tumors
- DNA from 10 benign neurofibromas
- DNA from Schwann cells of 6 healthy volunteers
I have to admit, I’m not a big fan of pooled sequencing. I like being able to match sequence to individual samples, by barcoding or other means. The good news is that the pool is rather small, so epigenetic changes unique to a single sample might be detectable, with sufficient coverage. The authors performed MeDIP and Illumina paired-end sequencing, generating ~65 million 2x50bp reads per pool. Some 72% could be uniquely mapped (map quality >=10) to the human genome by Maq. So far, so good.
Next, they normalized for copy number as assessed by Affy 6.0 arrays, to correct for potential bias in DNAm from copy number changes. Absolute DNAm values were inferred for 100bp windows using the BatMan algorithm, a Bayesian method for deconvoluting MeDIP-Seq data. On average, 67% of the ~26.7 million CpG sites in the haploid autosomal human reference were covered in each pool. The authors validated their DNAm data using two orthogonal approaches – BeadArray genotyping (Illumina Infinium) and bisulfite sequencing – with 77-78% overall concordance. These correlations are consistent with previous reports, and gave the authors the green light for genome-wide methylation analysis with the MeDIP-Seq data.
Comparative Analyses of DNA Methylation
BatMan produces a DNAm score quantifying the fraction of CpGs methylated in a given window. Using this value, regions were classified as low (<40%), intermediate (40-60%), or high (>60%) DNAm. First, the high-level view: there was only a 0.7% change in overall methylation between MPNSTs and the NF/SC controls. This was unexpected, as studies of other cancer types have shown a 10-20% reduction in methylation in tumors compared to normal cells. To dig deeper, the authors analyzed DNAm in the context of 16 types of genomic features (CpG islands and “shores”, regulatory regions, gene structures, and repeats). Most of these showed no significant difference in methylation between pools.
The most significant feature associated with DNAm changes comprised satellite repeats. Hypomethylation of repeats is a commonly cited feature of many cancers, but these regions have traditionally been refractory to DNAm assays, particularly array-based methods. Next-gen sequencing can often resolve these regions; indeed, the authors report “excellent coverage” of all repeat types using their MeDIP-Seq data. That’s impressive, considering the rather conservative mapping quality cutoff (10). Further, it allowed the authors to build global DNAm profiles of different repeat families in addition to gene and regulatory regions. The findings were very interesting:
- In contrast to previous studies, global hypomethylation of LINE repeats was unsupported. In fact, there was a slight decrease in low-DNAm regions among MPNSTs compared to controls overall, and L1/L2 repeats saw an increase in high-DNAm regions.
- SINE repeats showed a small (6%) decrease in high-DNAm regions in malignant tumors.
- Satellite repeats showed the starkest changes in DNAm, particularly between benign (NF) and malignant (MPNST) cell types.
Differentially Methylated Regions
Next, the authors performed pair-wise comparisons to identify differentially methylated regions (DMRs) between malignant, benign, and normal SC tissues. Each 1,000-bp window with average BatMan score differences of >= 33% in any pairwise comparison was called a DMR. This approach yielded 101,466 unique DMRs, of which 48% were hypermethylated and 52% hypomethylated in the tumor. Intriguingly, the malignant-benign comparison yielded the fewest DMRs, suggesting that these samples are more closely related to each other, from a methylation perspective, than they are to normal cells.
DMRs were mapped to the nearest genomic feature, and subjected to enrichment analysis. Interestingly, only 11% of DMRs were within 3kb of the nearest gene. Hypermethylated DMRs were enriched for non-Cpg-Island promoters, Cpg Island shores, LTRs, and LINES. Hypomethylated DMRs were enriched for non-CpG-Island promoters, SINEs, and satellite repeats. Neither type of DMR was significantly associated with CpG islands. This is striking, because most studies of differential DNAm to-date have focused on CpG islands. Apparently, the majority of DMRs fall outside of these regions. However, CpG island “shores” showed significant enrichment of both hyer- and hypo-methylation, emphasizing a key regulatory role for these sequences.
Genes Implicated by DMR
Some 3,690 genes were implicated in MPNST development and progression by this analysis. Consistent with previous studies, NF1 was not among them. However, there were some other interesting hits, including hypermethylation of the promoters of tumor suppressor genes MEST and WT1. There were some interesting candidate micro-RNA genes too, including miR-124, recently reported as a biomarker for high-grade cervical cancer. About 15% of known tissue-specific DMRs and cancer DMRs (which often coincide) were picked up by this study, including hypomethylated oncogenes YES1, MATK, E2F3, EGFR, and AFF1; as well as hypermethylated tumor suppressors TSC2, RUNX1, FOXD3, and RASSF1.
In summary, Feber et al have demonstrated the suitability of MeDIP-Seq for comparative methylome analyses in human cancers, and provided new insights into methylation’s role in progression from healthy/benign tissue to malignant tumors. I hope there’s more to come.
References
Feber A, Wilson GA, Zhang L, Presneau N, Idowu B, Down TA, Rakyan VK, Noon LA, Lloyd AC, Stupka E, Schiza V, Teschendorff AE, Schroth GP, Flanagan A, & Beck S (2011). Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome research PMID: 21324880
Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Gräf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Bäckdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavaré S, & Beck S (2008). A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nature biotechnology, 26 (7), 779-85 PMID: 18612301