At AGBT 2009 I presented a poster comparing 10 short read aligners on multiple sets of Illumina/Solexa sequencing data. This was an idea I’d conceived six months earlier, when it seemed that each new issue of Bioinformatics or Genome Research had an article about another short read alignment algorithm. We built Maq into the pipeline here over a year ago. It was a bit of a gamble to discard the Illumina-provided aligner (ELAND) and go with something else, but I think that the success of the AML cancer genome paper shows that our bet paid off. So as the list of alternative aligners grew, I came up with a set of questions to ask when considering any aligner for our pipeline here.
Ten Short Read Aligners I’ve Evaluated
For my AGBT poster I chose 10 aligners (including Maq) to install and evaluate on multiple Illumina/Solexa data sets:
| Aligner | Version | Developer | License |
| BFAST | 0.3.1 | UCLA | Academic |
| Bowtie | 0.9.8 | Salzberg/UMD | GNUPL |
| cross_match | 1.080721 | U. Wash. | Academic |
| CELL | 2.0 | CLC bio | Commercial |
| Maq | 0.7.1 | WTSI | GNUPL |
| Novoalign | 2.00.12 | Novocraft | Academic |
| RMAP | 0.41 | CSHL | Public |
| SeqMap | 1.0.12 | Stanford | Academic |
| Shrimp | 1.1.0 | U. Toronto | Public |
| SOAP | 2.01 | BGI | Academic |
How Should Alignment Tools Be Assessed?
When I’m considering an aligner for integration into our pipeline for high-throughput data processing (primarily Illumina/Solexa data), here are some questions I like to ask.
Usability and Documentation
Is the software easy to install/run? Is it available on Linux/OSX platforms?
Does it work without crashing? Does it handle problems gracefully?
Is there good documentation with example commands/workflows?
Is the algorithm described/published?
Features
Does it read native formats (Solexa, FASTA, FASTQ)?
Does it do paired-end alignment? Does it allow gaps in SE mode?
Does it include tools for mutation detection and coverage analysis?
Potential
What is the software license? Is the source code available?
Are developers working on the software? Are other groups actively using it?
Does it have read length/type limitations?
Trial 1: Simulated 36-bp Reads from C. elegans
The first data set to which I applied the alignment tools was created in silico using Maq’s simulation tools. First, I trained Maq-simulation on real data (36 bp Illumina PE library) from the YRI sample we’re sequencing for the 1000 Genomes Project. Then, I created a “mutated” version of the C. elegans genome that contained ~90,000 SNps and ~10,000 indels. From this mutated genome, using the training parameters, I told Maq to generate 1,000,000 simulated read-pairs (2m reads total). The key to this experiment is the read name, which tells us the true location in the genome that it came from:
| >chrI_7433167_7433425_1/1 TCGTATTATAACGACGAATACGCGTCAAGTGGGAGT >chrII_8451697_8451957_6086/1 CATATTTTTTAGAGGTTGGCGTCTCATGACACTCGT |
In essence, we have the “right” answer and can use it to determine if a read is placed correctly.
CPU Time to Align 1 Million Reads/Read-Pairs
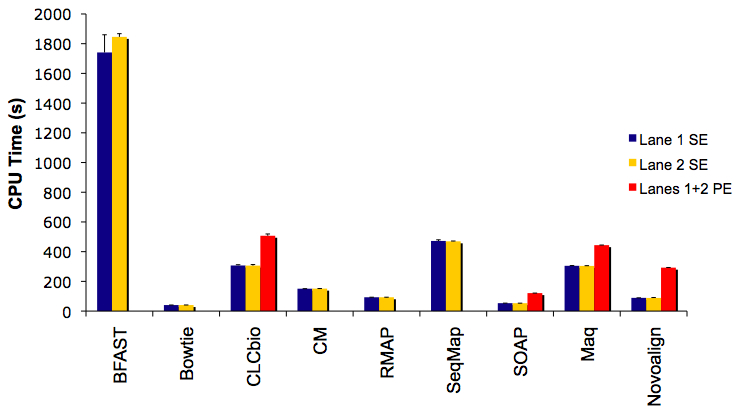
Not shown is SHRiMP, which took >3,300 seconds to complete. Even at this relatively small throughput, the speed advantage of Burrows-Wheeler Transformation indexers (Bowtie, SOAP) is clear.
Read Placement Results for 2m Reads
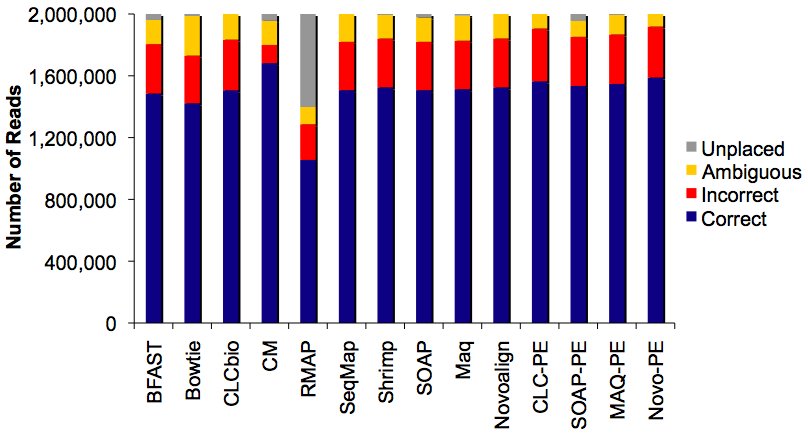
Because we know the true location of each read in the genome, we can tabulate the # of reads correctly aligned, incorrectly aligned, ambiguously aligned, or unplaced. RMAP was a disappointing outlier. The results were similar across the other aligners, with Novoalign paired-end and cross_match single-end placing the highest number of reads correctly.
Causes of Incorrectly Placed Reads
The fake mutations (SNPs) and indels were also recorded during this trial, and so next we sought to determine how often they caused read mis-alignment.
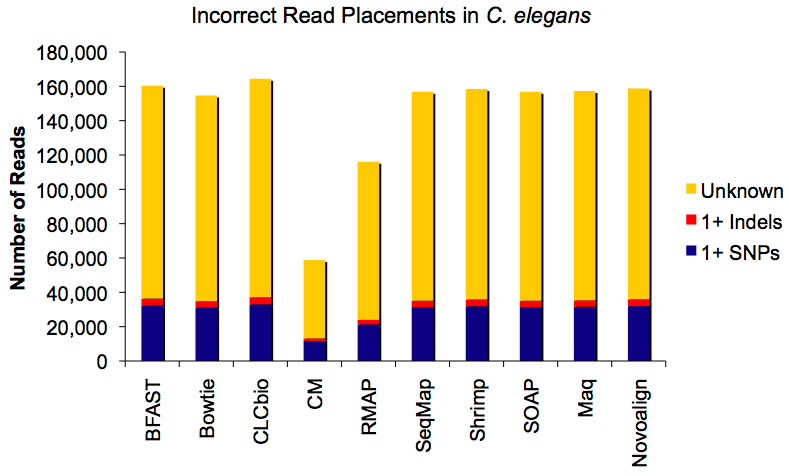
It seems that substitutions (SNPs) have a far greater influence than indels, but the higher effect tracks well with the excess of SNPs relative to indels (~90,000 vs 10,000). The bigger question: what’s causing the majority of mis-alignments (in yellow)? The simulation itself is a prime suspect, since Maq introduces errors (based on the training set read qualities) and variation (with a mutation rate of 0.001) during simulation, and these add to the mutations that I created in silico.
Trial 2: Real Data from a 36-bp Paired-End Illumina Run
The true test for short read aligners involves real data, particularly from large/complex mammalian genomes. To assess performance on this scale, we used whole-genome resequencing data from a Yoruban HapMap sample being sequenced as part of the 1000 Genomes Project. This was the real data set used to train the read simulations from Trial 1. It’s a 36-bp paired-end Illumina library, from which we took the first 1 million read pairs. Due to the disappointing speed of SHRiMP and diminished success of RMAP, these two aligners were dropped from future evaluations.
CPU Time to Align 1 Million Reads/Read-Pairs
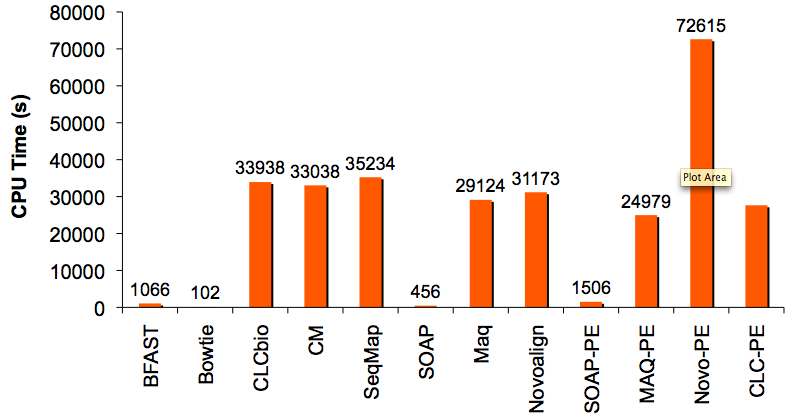
Aligning reads to the human genome is where remarkable differences in speed were observed. There were essentially two speed groups. The fastest were BWT-indexing aligners (Bowtie & SOAP) and BFAST, all of which finished in less than 20 minutes. The other aligners (CLCbio, CM, SeqMap, Maq, Novoalign) were comparable in speed to one another and completed in 8-9 hours. Notably, Novoalign’s paired-end mode at this scale was very slow (>20h), a weakness acknowledged to me privately by the authors.
Read Placement Results for ~2 Million Reads
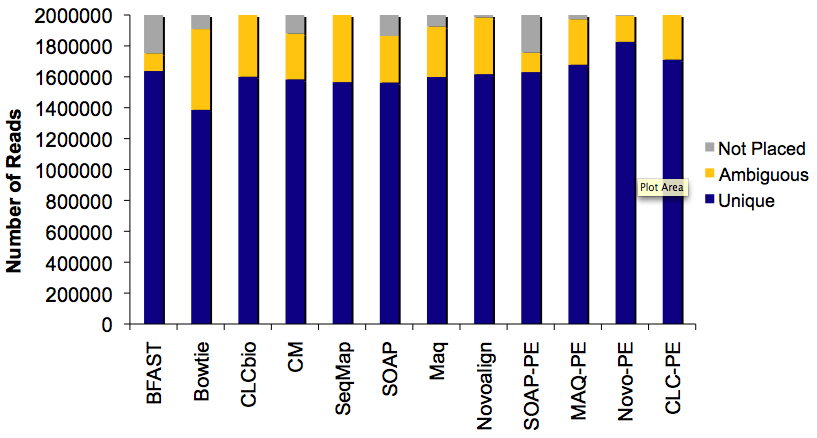
There were several interesting patterns in read placement between aligners. Most of them placed similar numbers of reads uniquely, yet the numbers of ambiguously-placed or unmapped reads varied widely. Novoalign in paired-end mode was the clear winner, though in fairness, it did take at least twice as long to complete as the other aligners.
Top Performing Short Read Aligners
Based on the performance comparisons, as well as less tangible results (like usability/interface, platform compatibility, aligner options/features, etc.), I’ve selected the best short read aligners in a few different ways.
| Alignment Need |
Recommendation | Notes |
| Proven Functionality | Maq | From what I heard at AGBT, nearly everyone uses Maq for aligning Illumina/Solexa data. It’s clearly the most widely used and probably the most publication-proven algorithm. |
| Speed | Bowtie, SOAP | The Burrows-Wheeler Transformation indexing aligners have a clear speed advantage. SOAP has the added benefit of paired-end mode, something that Bowtie developer Ben Langmead promises soon. |
| Sensitivity | Novoalign, cross_match | Both of these aligners have algorithmic advantages over programs like Maq. Novoalign allows gaps in single-end mode, and seems better suited to handling mismatches as well. The cross_match program, while poorly documented, had the best overall sensitivity in my testing. |
| Usability | CLC bio, SeqMap | For users with little or no bio-informatics experience, I have two recommendations. If you have money to spend, consider CLC bio’s CELL2 suite, which includes many easy-to-use programs. On the cheap, SeqMap was also an all-around user-friendly program in terms of inputs and outputs. |
| Flexibility | BFAST | It’s fair to say that I still don’t understand all of BFAST’s capabilities. You can generate numerous indexes of a reference genome to be iteratively searched, where each one allows different levels/types of variation in the reads. |
| Variant Detection | SOAP, Novoalign | Novoalign makes the list because of added sensitivity to substitutions and gaps, which makes it *ideal* for mining Maq-unplaced reads for variation. SOAP developers at BGI have already released a SNP caller (SOAPsnp) and promise an indel caller too. |
Further slides from my AGBT poster will be added as time allows. For now, I’m working on getting my baby to sleep through the night.
